| DPOGS200932 | ||
|---|---|---|
| Transcript | DPOGS200932-TA | 630 bp |
| Protein | DPOGS200932-PA | 209 aa |
| Genomic position | DPSCF300301 + 144574-145543 | |
| RNAseq coverage | 376x (Rank: top 32%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL002514 | 3e-101 | 96.17% | |
| Bombyx | BGIBMGA000303-TA | 3e-101 | 95.69% | |
| Drosophila | CG8500-PA | 1e-86 | 75.57% | |
| EBI UniRef50 | UniRef50_Q96HU8 | 3e-83 | 71.77% | GTP-binding protein Di-Ras2 n=90 Tax=Bilateria RepID=DIRA2_HUMAN |
| NCBI RefSeq | XP_001953593.1 | 3e-86 | 76.92% | GF17842 [Drosophila ananassae] |
| NCBI nr blastp | gi|194742199 | 6e-85 | 76.92% | GF17842 [Drosophila ananassae] |
| NCBI nr blastx | gi|194742199 | 9e-90 | 78.73% | GF17842 [Drosophila ananassae] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0016020 | 1.3e-128 | membrane | |
| GO:0007165 | 1.3e-128 | signal transduction | ||
| GO:0006184 | 1.3e-128 | GTP catabolic process | ||
| GO:0005525 | 1.3e-128 | GTP binding | ||
| GO:0003924 | 1.3e-128 | GTPase activity | ||
| GO:0007264 | 1.1e-45 | small GTPase mediated signal transduction | ||
| GO:0015031 | 6.4e-23 | protein transport | ||
| GO:0005622 | 1e-11 | intracellular | ||
| KEGG pathway | ||||
| InterPro domain | [1-190] IPR020849 | 1.3e-128 | Small GTPase superfamily, Ras type | |
| [9-169] IPR001806 | 1.1e-45 | Small GTPase superfamily | ||
| [8-174] IPR003579 | 6.4e-23 | Small GTPase superfamily, Rab type | ||
| [6-167] IPR005225 | 1.1e-21 | Small GTP-binding protein domain | ||
| [10-170] IPR003578 | 1e-11 | Small GTPase superfamily, Rho type | ||
| Orthology group | MCL14299 | Single-copy universal gene | ||
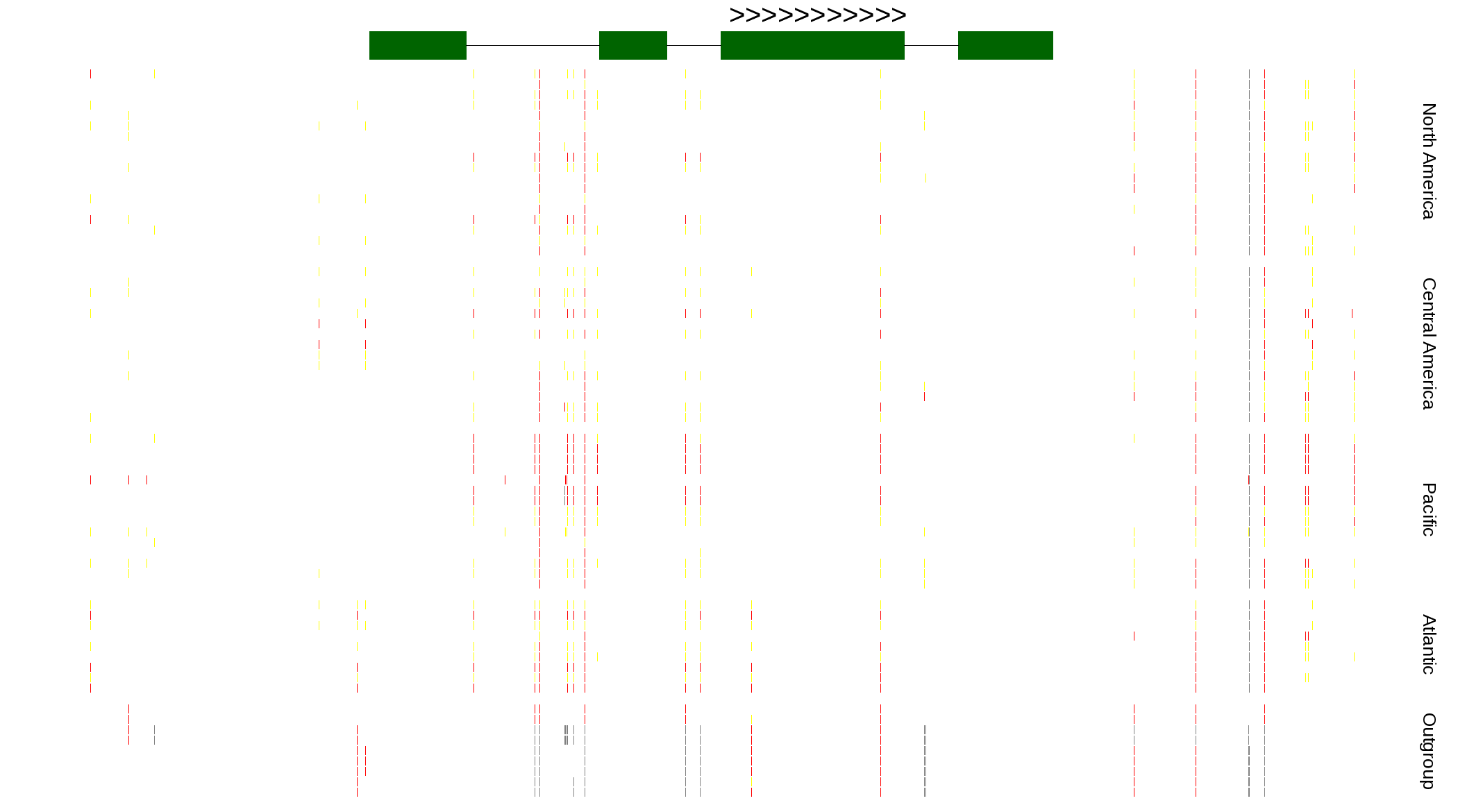
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS200932-TA
ATGCCGGAGCAAAGCAATGATTACCGTGTTGTTGTATTTGGTGCAGGCGGTGTTGGTAAAAGTTCGTTAGTGTTGAGATTTGTTAAAGGTACCTTCAGGGAATCATATATTCCAACGATTGAAGATACTTACAGACAAGTCATAAGCAGCAACAAAACTATATGCACTCTTCAGATAACTGACACAACAGGGTCACACCAGTTTCCAGCAATGCAAAGGTTATCTATTAGTAAGGGGCATGCTTTCATACTGGTGTATTCAGTTAGCAGTCGGCAATCGTTGGAGGAATTGAAGCCAATATGGCAGACAATAAAAGAGATTAAAGGTGCAGAATTACCAAATATACCTGTCATGTTGGCTGGTAACAAGTGTGATGAGACTCCTGAGATACGGGAAGTATCAGCTGCCGAGGGTCAGGCTCAGGCTCAAAATTGGGGAGTGTCATTCATGGAAACTTCCGCGAAAACTAATCATAATGTCACACAGCTTTTCCAGGAGCTGCTTAACATGGAAAAGAATAGAAATGTGTCACTTCAAGTTGATGGAAAGGGTAAGAAGAAAAAGGATAAAGTTGTTAAAGATGGTGGTGAACCATCGGCTAGTGGTAGAGAGAAATGCCTTGTCATGTAA
>DPOGS200932-PA
MPEQSNDYRVVVFGAGGVGKSSLVLRFVKGTFRESYIPTIEDTYRQVISSNKTICTLQITDTTGSHQFPAMQRLSISKGHAFILVYSVSSRQSLEELKPIWQTIKEIKGAELPNIPVMLAGNKCDETPEIREVSAAEGQAQAQNWGVSFMETSAKTNHNVTQLFQELLNMEKNRNVSLQVDGKGKKKKDKVVKDGGEPSASGREKCLVM-