| DPOGS201659 | ||
|---|---|---|
| Transcript | DPOGS201659-TA | 351 bp |
| Protein | DPOGS201659-PA | 116 aa |
| Genomic position | DPSCF300103 - 519964-521190 | |
| RNAseq coverage | 865x (Rank: top 15%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL011925 | 4e-63 | 97.41% | |
| Bombyx | BGIBMGA005413-TA | 1e-24 | 94.34% | |
| Drosophila | spt4-PA | 2e-50 | 73.28% | |
| EBI UniRef50 | UniRef50_Q9TVQ5 | 4e-48 | 73.28% | Transcription elongation factor SPT4 n=34 Tax=Opisthokonta RepID=SPT4H_DROME |
| NCBI RefSeq | XP_624457.1 | 8e-59 | 89.66% | PREDICTED: similar to spt4 CG12372-PA isoform 2 [Apis mellifera] |
| NCBI nr blastp | gi|307211608 | 8e-58 | 91.38% | Transcription elongation factor SPT4 [Harpegnathos saltator] |
| NCBI nr blastx | gi|307211608 | 6e-59 | 91.38% | Transcription elongation factor SPT4 [Harpegnathos saltator] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0032786 | 3.5e-71 | positive regulation of transcription elongation, DNA-dependent | |
| GO:0005634 | 3.5e-71 | nucleus | ||
| GO:0006355 | 3.5e-71 | regulation of transcription, DNA-dependent | ||
| GO:0008270 | 3.5e-71 | zinc ion binding | ||
| KEGG pathway | ||||
| InterPro domain | [1-117] IPR016046 | 2.6e-78 | Transcription initiation Spt4-like | |
| [1-117] IPR009287 | 3.5e-71 | Transcription initiation Spt4 | ||
| [13-90] IPR022800 | 2.4e-32 | Spt4/RpoE2 zinc finger | ||
| Orthology group | MCL13540 | Single-copy universal gene | ||
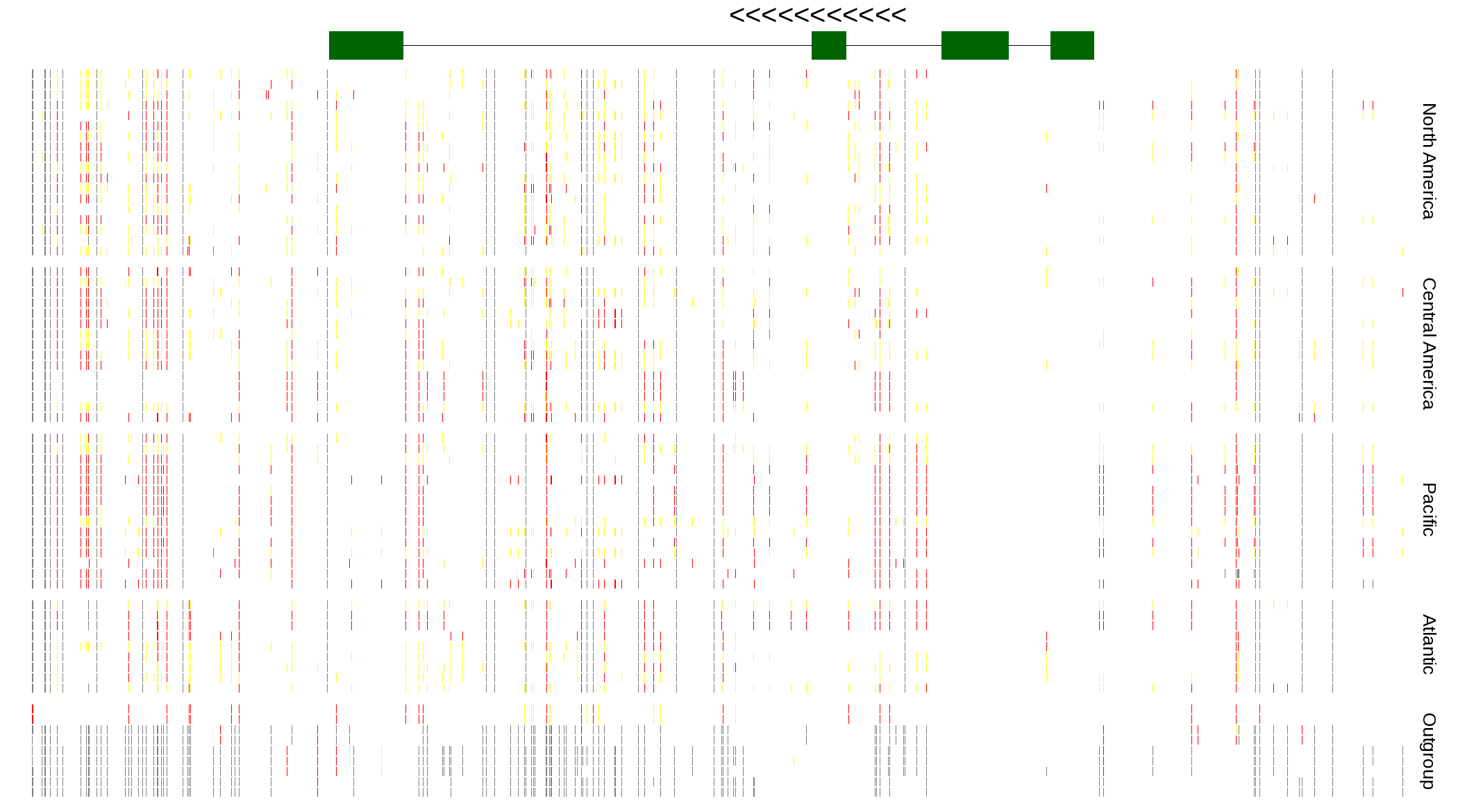
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS201659-TA
ATGTCCCTCGACACGGTTCCAAAAGATTTAAGAGGTTTAAGAGCATGTTTAGTTTGTTCTCTAATTAAGACATTCGATCAGTTCGAATATGATGGCTGTGACAATTGCGATGAGTTTCTCCGGATGAAGAACAATAAGGACAATGTATATGACTGCACAAGTAACAATTTTGATGGAATGATTGCAGTTATGAGCCCTGTTGATAGTTGGGTCTGCAAGTGGCAGAGAATAAGTAGATTTTGTCAAGGTGTGTATGCTATATCTGTGTCGGGCCGGCTTCCAGCTGGTGTTATAAGAGAAATGAAGAGTCGCGGTATCGCCTACAGACCCAGAGACACGAGCCAGAGATAA
>DPOGS201659-PA
MSLDTVPKDLRGLRACLVCSLIKTFDQFEYDGCDNCDEFLRMKNNKDNVYDCTSNNFDGMIAVMSPVDSWVCKWQRISRFCQGVYAISVSGRLPAGVIREMKSRGIAYRPRDTSQR-