| DPOGS201825 | ||
|---|---|---|
| Transcript | DPOGS201825-TA | 567 bp |
| Protein | DPOGS201825-PA | 188 aa |
| Genomic position | DPSCF300482 - 29391-39097 | |
| RNAseq coverage | 19x (Rank: top 80%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL011198 | 2e-38 | 95.79% | |
| Bombyx | BGIBMGA013336-TA | 8e-11 | 90.20% | |
| Drosophila | Dgk-PE | 9e-17 | 34.38% | |
| EBI UniRef50 | UniRef50_F4WSF1 | 2e-34 | 50.59% | Diacylglycerol kinase beta n=17 Tax=Coelomata RepID=F4WSF1_ACREC |
| NCBI RefSeq | XP_974283.2 | 2e-33 | 49.44% | PREDICTED: similar to diacylglycerol kinase, beta 90kDa [Tribolium castaneum] |
| NCBI nr blastp | gi|328792207 | 9e-38 | 52.94% | PREDICTED: diacylglycerol kinase 1 [Apis mellifera] |
| NCBI nr blastx | gi|380027152 | 4e-37 | 52.94% | PREDICTED: diacylglycerol kinase 1-like [Apis florea] |
| Group | ||||
|---|---|---|---|---|
| KEGG pathway | tca:663130 | 6e-33 | ||
| K00901 (E2.7.1.107, DGK, dgkA) | maps-> | Phosphatidylinositol signaling system | ||
| Glycerolipid metabolism | ||||
| Glycerophospholipid metabolism | ||||
| Orthology group | MCL19803 | Insect specific | ||
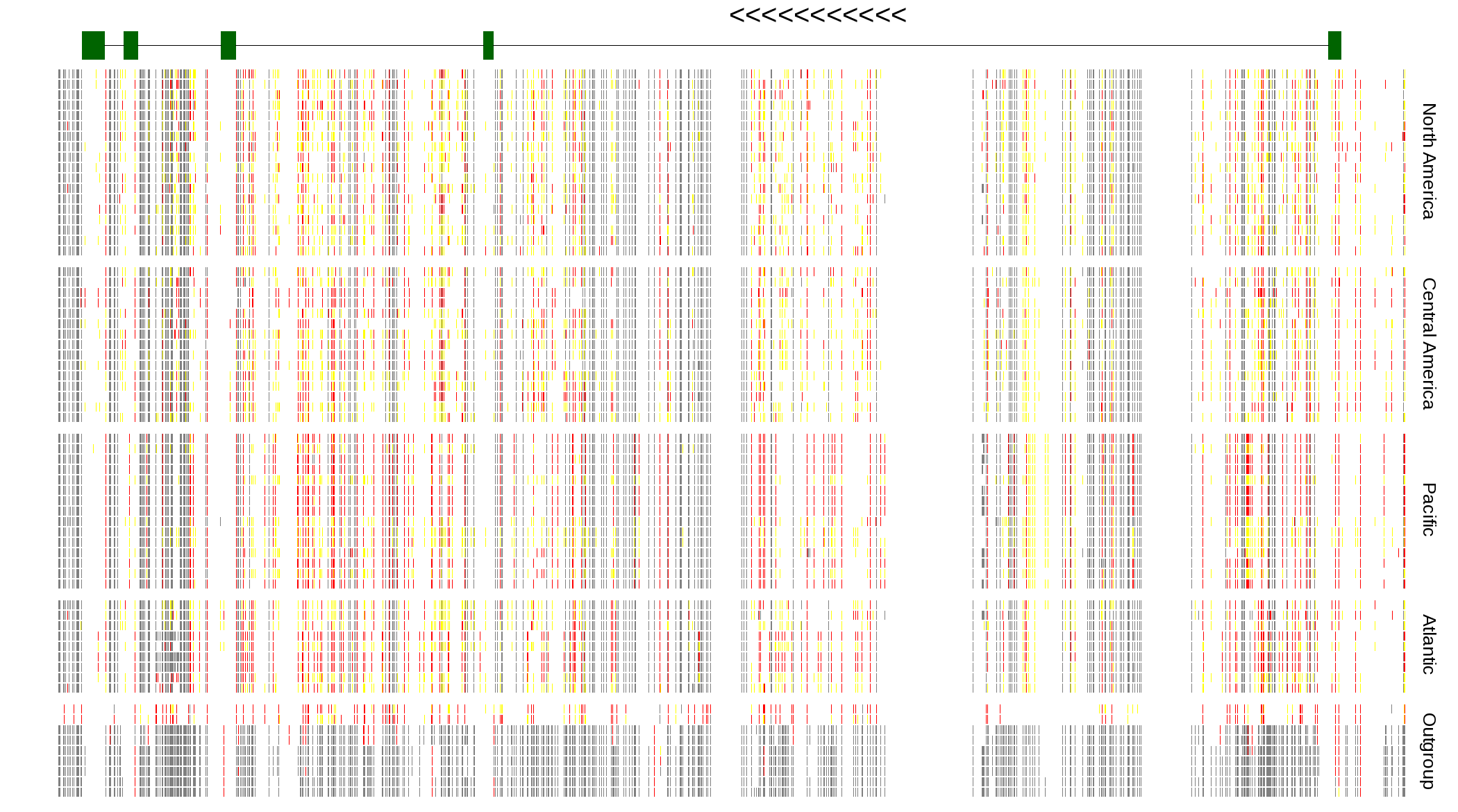
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS201825-TA
ATGAGAATGGCTGCGATGCAAAATGATCCGTATGCTGACAACGTAGAGCACGAATTCGGAATCGGCGTGACGTTTCCAAGGATTGTTGACCCAGTGGACATTGACTTCGAAGGCTTCCGATGGTTCCTGGACACTTTCCTCGAGGTCACCACCCCAGATGACCTGGCCAGGCATCTGTTTCTGTCCTTCGTGCGAAGAGACAAGAGACCAGCTCTACGTGAAATGGCAGCAGCTAGTTCGACCACCGCTTGTGCTGCCGTCACTGCTCATGCTGACCATCCGCCAAAATTAAGTCTGGCCTCCAAGATCCATGGACTCGCGGAGAGATTGCAGCAGCTTGGAAAGAGTTCAGGAGATTCGGTGGATGGAGAGAAGGGTTCTAGAAGCAGAACTGGCAGTGTTCATCCAATGTTCTCTGTGACGACTCATCCATCATACTCCTCCCACGAGCTGTGTCTGGGGAGGTTGGAGACCAGCCCCAGTCACAGCCAAATGTCGAGGAACTCCAGCAGGAAAAGCAGCAATAGCTTACGTGTGAATCAGTCAACGAAGATAGAAGGTACATGA
>DPOGS201825-PA
MRMAAMQNDPYADNVEHEFGIGVTFPRIVDPVDIDFEGFRWFLDTFLEVTTPDDLARHLFLSFVRRDKRPALREMAAASSTTACAAVTAHADHPPKLSLASKIHGLAERLQQLGKSSGDSVDGEKGSRSRTGSVHPMFSVTTHPSYSSHELCLGRLETSPSHSQMSRNSSRKSSNSLRVNQSTKIEGT-