| DPOGS201917 | ||
|---|---|---|
| Transcript | DPOGS201917-TA | 750 bp |
| Protein | DPOGS201917-PA | 249 aa |
| Genomic position | DPSCF300507 + 43712-45237 | |
| RNAseq coverage | 332x (Rank: top 35%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL005550 | 1e-108 | 79.92% | |
| Bombyx | BGIBMGA004218-TA | 2e-120 | 85.25% | |
| Drosophila | CG10166-PA | 3e-112 | 79.08% | |
| EBI UniRef50 | UniRef50_Q9WU83 | 1e-110 | 77.41% | Dolichol-phosphate mannosyltransferase n=92 Tax=Eukaryota RepID=DPM1_CRIGR |
| NCBI RefSeq | XP_392640.1 | 1e-119 | 84.65% | PREDICTED: similar to Probable dolichol-phosphate mannosyltransferase (Dolichol-phosphate mannose synthase) (Dolichyl-phosphate beta-D-mannosyltransferase) (Mannose-P-dolichol synthase) (MPD synthase) (DPM synthase) [Apis mellifera] |
| NCBI nr blastp | gi|380011871 | 1e-118 | 85.06% | PREDICTED: probable dolichol-phosphate mannosyltransferase-like [Apis florea] |
| NCBI nr blastx | gi|380011871 | 2e-116 | 85.06% | PREDICTED: probable dolichol-phosphate mannosyltransferase-like [Apis florea] |
| Group | ||||
|---|---|---|---|---|
| KEGG pathway | ame:409115 | 3e-119 | ||
| K00721 (DPM1) | maps-> | N-Glycan biosynthesis | ||
| InterPro domain | [17-187] IPR001173 | 8e-37 | Glycosyl transferase, family 2 | |
| Orthology group | MCL13459 | Single-copy universal gene | ||
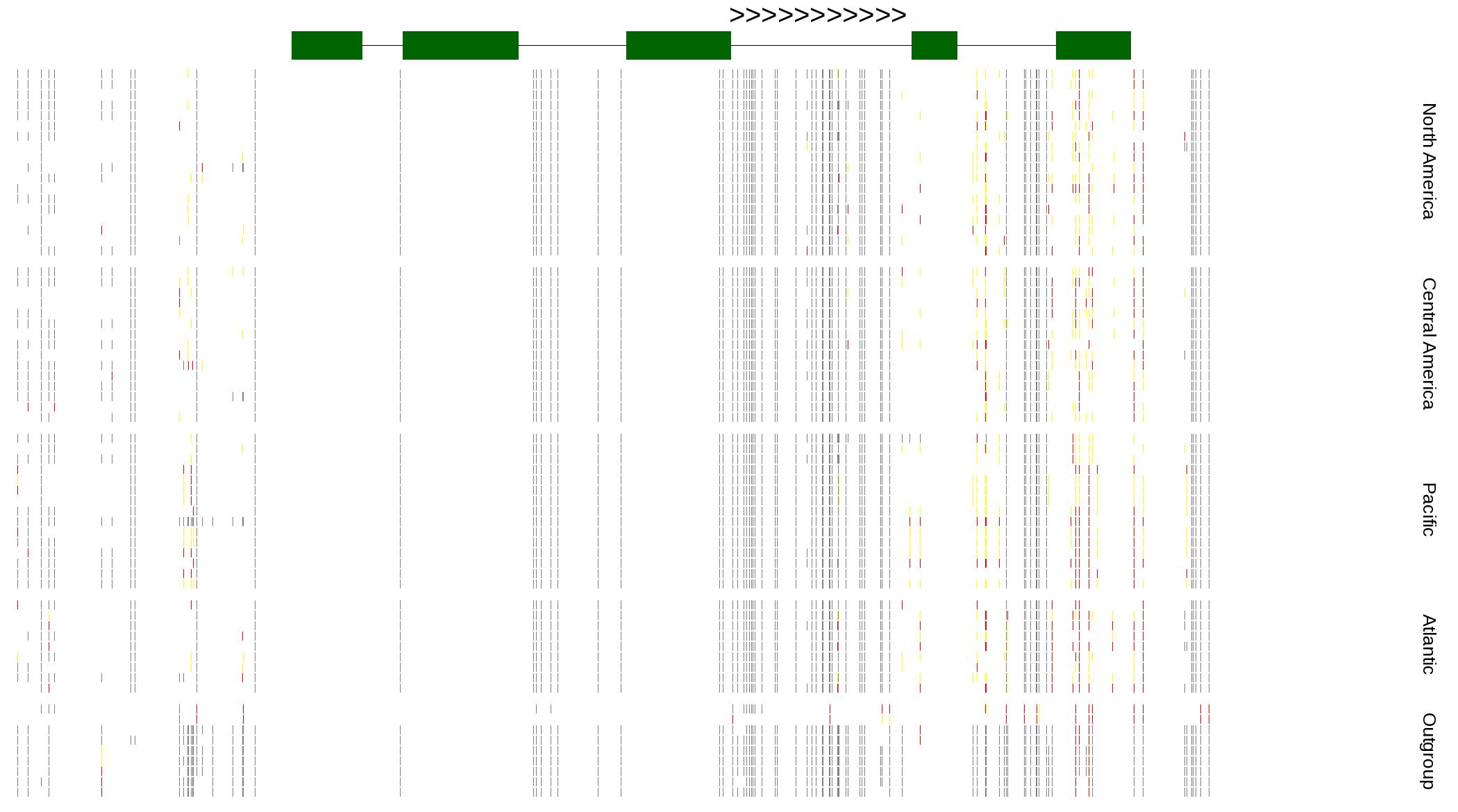
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS201917-TA
ATGGCGGCCACGGTTTCAGATTCAGAAATCACAAAAAGTGATAAGTATTCAATTTTACTTCCAACCTACAACGAAAGAGAGAATTTACCGATCATAATATGGCTCATTATTAAGTATCTAGACAATAGTGGACACGATTACGAAGTGATTGTGATTGATGATGGAAGTCCCGATGGCACTTTGGAAGTGGCTAAGCAGCTACAGAAATTATACGGGTCGGATAAAATAGTTTTAAGACCTCGGGAAAAGAAATTAGGTTTAGGCACCGCATACATACATGGTATAAAACATGCTACGGGTAATTTTATCATAATTATGGACGCGGATTTAAGTCACCACCCAAAATTCATACCAAATTTCATTGAGTTACAAAAGAAACACGATCTAGATGTGGTTTCTGGGACTCGTTACAAGGATGGTGGTGGAGTGTATGGTTGGGACTTCAAAAGGAAATTAATATCCAGGGGTGCCAACTTTATAACACAGTTGCTTCTGAGGCCCGGTGCATCAGACTTGACGGGATCCTTTAGACTGTATAAGAAAGATATTCTTCAAAAATTAATTGACAGCTGTGTGTCTAAGGGCTATGTCTTCCAAATGGAAATGATTATAAGAGCAAGACAGCTAGAATACACTATAGGCGAGGTGCCGATTACGTTTGTTGATCGTGTGTACGGAGTTTCCAAGCTCGGAGGATCGGAGATTATTCAGTTTGCGAAGGCTTTACTTTATTTGTTTGCCACCACATAA
>DPOGS201917-PA
MAATVSDSEITKSDKYSILLPTYNERENLPIIIWLIIKYLDNSGHDYEVIVIDDGSPDGTLEVAKQLQKLYGSDKIVLRPREKKLGLGTAYIHGIKHATGNFIIIMDADLSHHPKFIPNFIELQKKHDLDVVSGTRYKDGGGVYGWDFKRKLISRGANFITQLLLRPGASDLTGSFRLYKKDILQKLIDSCVSKGYVFQMEMIIRARQLEYTIGEVPITFVDRVYGVSKLGGSEIIQFAKALLYLFATT-