| DPOGS202445 | ||
|---|---|---|
| Transcript | DPOGS202445-TA | 459 bp |
| Protein | DPOGS202445-PA | 152 aa |
| Genomic position | DPSCF300174 - 300381-302408 | |
| RNAseq coverage | 135x (Rank: top 56%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL016404 | 8e-58 | 69.01% | |
| Bombyx | BGIBMGA009966-TA | 2e-60 | 71.83% | |
| Drosophila | CG3835-PB | 1e-39 | 50.35% | |
| EBI UniRef50 | UniRef50_D6WE94 | 4e-48 | 58.16% | Putative uncharacterized protein n=2 Tax=Tribolium castaneum RepID=D6WE94_TRICA |
| NCBI RefSeq | XP_001807867.1 | 7e-49 | 58.16% | PREDICTED: similar to d-lactate dehydrognease 2, putative [Tribolium castaneum] |
| NCBI nr blastp | gi|189235632 | 1e-47 | 58.16% | PREDICTED: similar to d-lactate dehydrognease 2, putative [Tribolium castaneum] |
| NCBI nr blastx | gi|270003457 | 1e-46 | 58.16% | hypothetical protein TcasGA2_TC002688 [Tribolium castaneum] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0050660 | 6.2e-38 | flavin adenine dinucleotide binding | |
| GO:0003824 | 6.2e-38 | catalytic activity | ||
| KEGG pathway | mcc:702028 | 1e-39 | ||
| K00100 (E1.1.1.-) | maps-> | Linoleic acid metabolism | ||
| Bisphenol A degradation | ||||
| Fructose and mannose metabolism | ||||
| Butanoate metabolism | ||||
| Tetrachloroethene degradation | ||||
| InterPro domain | [9-144] IPR004113 | 6.2e-38 | FAD-linked oxidase, C-terminal | |
| [9-147] IPR016164 | 1.6e-34 | FAD-linked oxidase-like, C-terminal | ||
| [105-145] IPR016171 | 1.8e-10 | Vanillyl-alcohol oxidase, C-terminal subdomain 2 | ||
| Orthology group | ||||
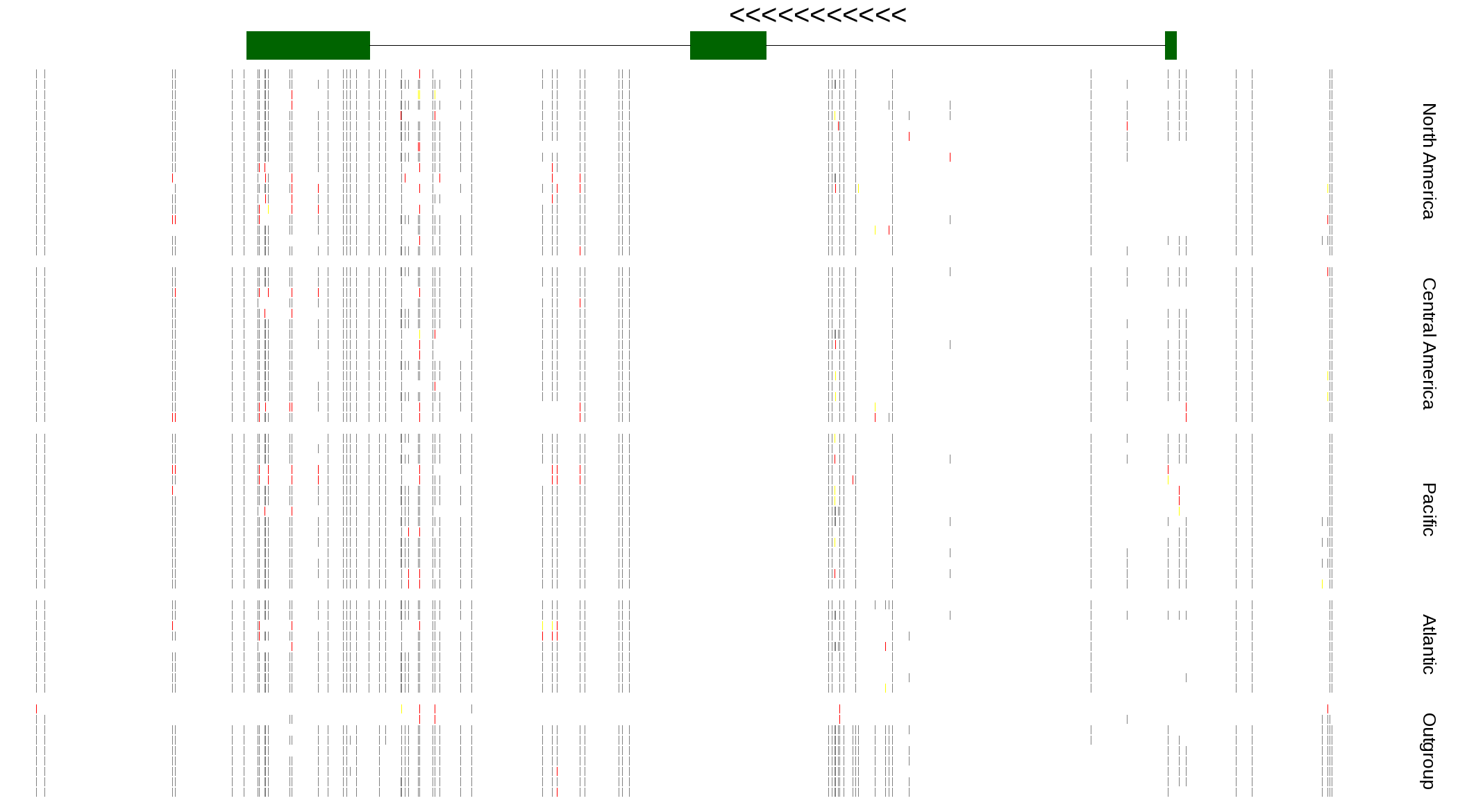
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS202445-TA
ATGCAAACGTTTTTCAAACTCATGGCGATCTGGAACATTCGGGAGAGCATCGCGGGTTCTGGTTTGATTGACGGCTATGTATTCAAATACGACGTGTCCCTCCCGCTGAGAAACTACTACGATCTCGTGGAAGACCTTCGAAAGAAGCTAGGAGACCGCGTCACCAGAGTTTACGGATACGGACATGTTGGTGACGGCAATATTCATATAAATGTAACAGTGCCGCAATATTCAAAGGAAATTAATTCTGAATTGGAGCCGTATATTTTCGAACAAGTATCCAAACTGAAGGGTTCTATCAGCGCCGAGCACGGCGTTGGTTTTAGGAAACCACAGTTCATACATTACAGTCAAGGAGACACTTCCTTGCAGCTAATGAGGGATTTAAAAAGAACCATGGATCCCAACGGCATACTGAACCCATACAAAGTACTGCCGGATGTTGCCAGTCATGAATAA
>DPOGS202445-PA
MQTFFKLMAIWNIRESIAGSGLIDGYVFKYDVSLPLRNYYDLVEDLRKKLGDRVTRVYGYGHVGDGNIHINVTVPQYSKEINSELEPYIFEQVSKLKGSISAEHGVGFRKPQFIHYSQGDTSLQLMRDLKRTMDPNGILNPYKVLPDVASHE-