| DPOGS202844 | ||
|---|---|---|
| Transcript | DPOGS202844-TA | 723 bp |
| Protein | DPOGS202844-PA | 240 aa |
| Genomic position | DPSCF300018 + 1007519-1008487 | |
| RNAseq coverage | 1009x (Rank: top 13%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL009301 | 3e-73 | 60.83% | |
| Bombyx | BGIBMGA010485-TA | 4e-104 | 77.50% | |
| Drosophila | aph-1-PA | 4e-68 | 54.27% | |
| EBI UniRef50 | UniRef50_Q9VQG2 | 5e-66 | 54.27% | Gamma-secretase subunit Aph-1 n=15 Tax=Endopterygota RepID=APH1_DROME |
| NCBI RefSeq | XP_392345.2 | 7e-72 | 56.96% | PREDICTED: similar to anterior pharynx defective 1 CG2855-PA [Apis mellifera] |
| NCBI nr blastp | gi|380011221 | 6e-72 | 56.36% | PREDICTED: gamma-secretase subunit Aph-1-like [Apis florea] |
| NCBI nr blastx | gi|380011221 | 6e-76 | 56.36% | PREDICTED: gamma-secretase subunit Aph-1-like [Apis florea] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0016485 | 8.9e-93 | protein processing | |
| GO:0016021 | 8.9e-93 | integral to membrane | ||
| GO:0043085 | 8.9e-93 | positive regulation of catalytic activity | ||
| KEGG pathway | ame:408814 | 2e-71 | ||
| K06172 (APH1) | maps-> | Alzheimer's disease | ||
| Notch signaling pathway | ||||
| InterPro domain | [2-229] IPR009294 | 8.9e-93 | Aph-1 | |
| Orthology group | MCL11051 | Multiple-copy universal gene | ||
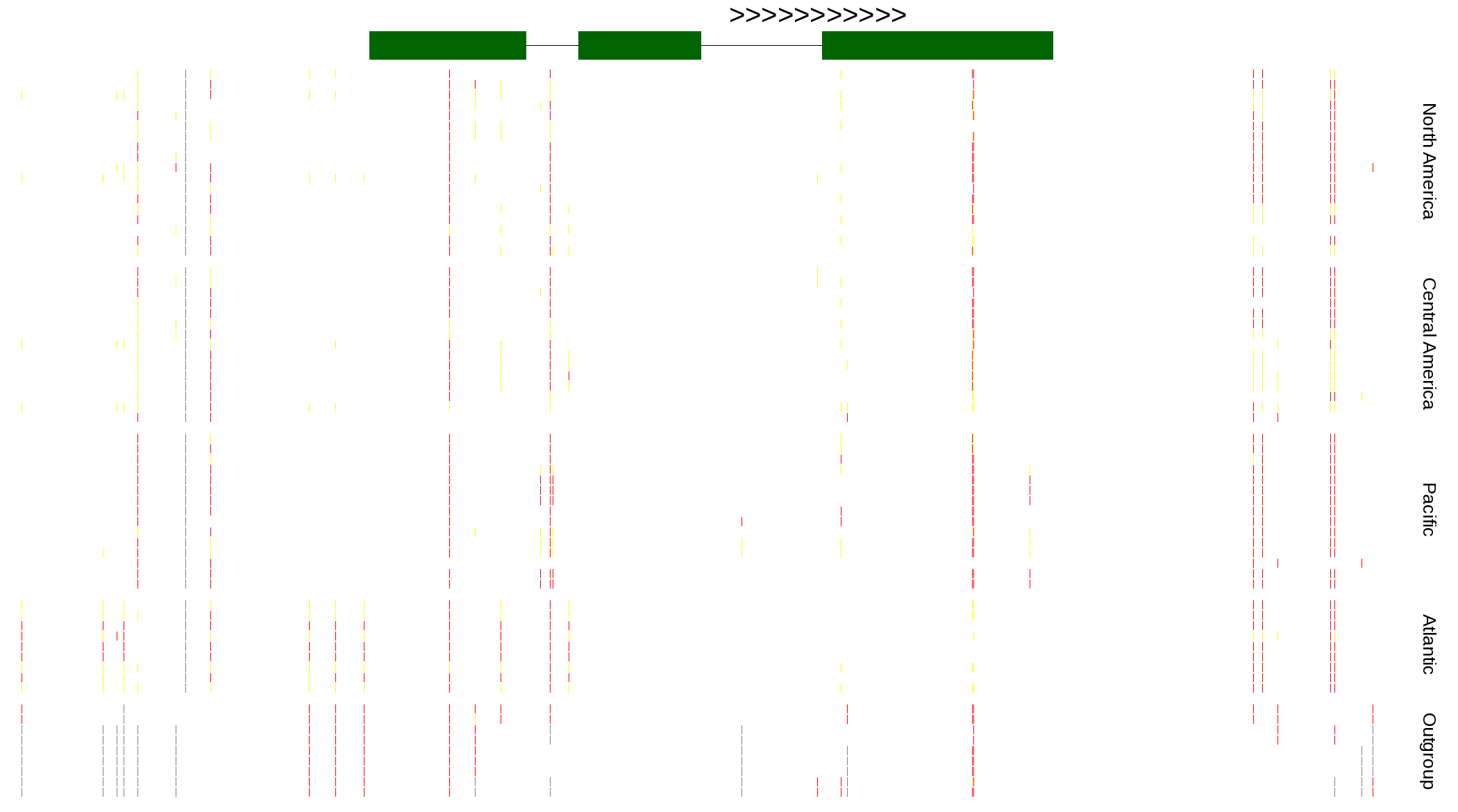
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS202844-TA
ATGACTGCCGCAGAGTTTTTCAGTTGTTCTCTTCTAGCCTTCGGAGCGCCTTTGGCTATGTTCGCTTTAACGATAGCCAACGACCCTGTGCGGATCATAATCATGATAGCTGCCGCGTTTTTTTGGCTGCTATCATTTCTACTGTCTTCGATGCTGTGGTACGCCGTGGTACCCCTACGGTCCTATTTAGCTTTTGGAATGGTTTTCGCGATATTAATCCAGGAGACTTTTCGCTTTGGTGTGTATCTTCTATTGAAAAGGACTGAATCAGGATTGAAGGAAATATCAGAAAACCATGAGATTGGTAGTAATAAGGCTGAGATTGCTTATGTGTCAGGCTTTGGGTTCGGAGCCATGAGTGGGGCATTTGCAATCATTAATGTGTTGGCAGATTCTATTGGTCCTGGAACGTTAGGACTTCGTCATGGAACGGAATACTTCTTTGTTACGAGTGCGTCTATGACTCTCTGCATGACGCTGCTCAACACATTTTGGACCGTCATCTTCTTTAATGGACTGGACAATAAGAGCTACTTGAAGATCCTGTGGGTGATCGGCTCACATTTTCTAGTGTCCGGATTGAGTTTACTCAATAGGAAAGAACTGTATGTGGCTACAATAATCCCATCATACATAATTCTTGTAATAACTCTATATATATCATTCAAGTGCGCCGGCGGTTCTAGTAAAGGTTTTCTGCAGACATTCCGGGCTAGAACTTAA
>DPOGS202844-PA
MTAAEFFSCSLLAFGAPLAMFALTIANDPVRIIIMIAAAFFWLLSFLLSSMLWYAVVPLRSYLAFGMVFAILIQETFRFGVYLLLKRTESGLKEISENHEIGSNKAEIAYVSGFGFGAMSGAFAIINVLADSIGPGTLGLRHGTEYFFVTSASMTLCMTLLNTFWTVIFFNGLDNKSYLKILWVIGSHFLVSGLSLLNRKELYVATIIPSYIILVITLYISFKCAGGSSKGFLQTFRART-