| DPOGS202970 | ||
|---|---|---|
| Transcript | DPOGS202970-TA | 399 bp |
| Protein | DPOGS202970-PA | 132 aa |
| Genomic position | DPSCF300259 - 168310-168801 | |
| RNAseq coverage | 11x (Rank: top 83%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL006029 | 4e-33 | 65.62% | |
| Bombyx | BGIBMGA005191-TA | 2e-42 | 60.83% | |
| Drosophila | Mdh2-PA | 2e-06 | 41.43% | |
| EBI UniRef50 | UniRef50_E0VRQ6 | 2e-08 | 32.81% | Malate dehydrogenase, putative n=1 Tax=Pediculus humanus corporis RepID=E0VRQ6_PEDHC |
| NCBI RefSeq | XP_002428800.1 | 3e-09 | 32.81% | malate dehydrogenase, putative [Pediculus humanus corporis] |
| NCBI nr blastp | gi|242016382 | 7e-08 | 32.81% | malate dehydrogenase, putative [Pediculus humanus corporis] |
| NCBI nr blastx | gi|242016382 | 2e-07 | 32.81% | malate dehydrogenase, putative [Pediculus humanus corporis] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0055114 | 4e-09 | oxidation-reduction process | |
| GO:0016491 | 4e-09 | oxidoreductase activity | ||
| GO:0005488 | 5.2e-09 | binding | ||
| KEGG pathway | pop:POPTR_822907 | 5e-07 | ||
| K00026 (MDH2) | maps-> | Citrate cycle (TCA cycle) | ||
| Pyruvate metabolism | ||||
| Carbon fixation in photosynthetic organisms | ||||
| Glyoxylate and dicarboxylate metabolism | ||||
| InterPro domain | [51-112] IPR001236 | 4e-09 | Lactate/malate dehydrogenase, N-terminal | |
| [51-117] IPR016040 | 5.2e-09 | NAD(P)-binding domain | ||
| Orthology group | MCL30314 | Lepidoptera specific | ||
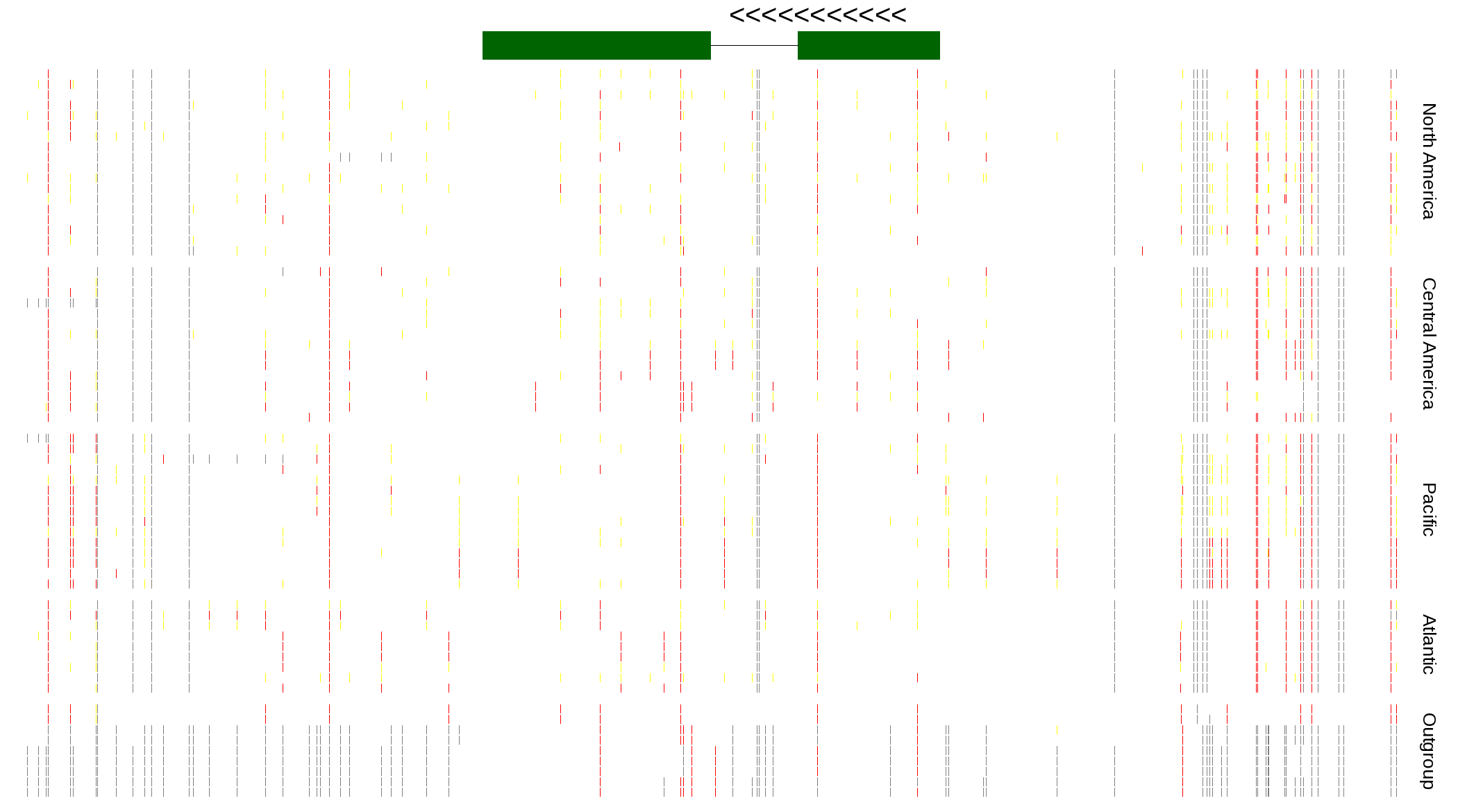
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS202970-TA
ATGTTCCGACAAATGACGCTCCTCCTGAGATCAAACAATGTCTCATCGCAAACTATAGCTAAATACCAGAATTATGTAATGTCCGAGGCTTTGGCGCATCTCGAACGGGAATGTGAGAAATACTGTCCAGTCCCAAGATTTCCACCTAAAAAAGTGACCGTTGTCGGCGCTGGTAGCGACGTCGGCTGTGTTGCCTCACTGTTCCTTAAACAGGAAAAGCTTATAAAGACACTGGCTTTATATGATGATGACCCAAATAACAAAGTACTTGGCATGGCTACCGACGTGGCTCACATAGACACCAGCACAGAGGTAGAAGCGTATCAAGGAAGAGCCTACCTAAAGACAGCCCTATGGGTACTGAAATATAAAATAATAATATTACACTACGAGAATTAA
>DPOGS202970-PA
MFRQMTLLLRSNNVSSQTIAKYQNYVMSEALAHLERECEKYCPVPRFPPKKVTVVGAGSDVGCVASLFLKQEKLIKTLALYDDDPNNKVLGMATDVAHIDTSTEVEAYQGRAYLKTALWVLKYKIIILHYEN-