| DPOGS202972 | ||
|---|---|---|
| Transcript | DPOGS202972-TA | 546 bp |
| Protein | DPOGS202972-PA | 181 aa |
| Genomic position | DPSCF300259 + 277979-282123 | |
| RNAseq coverage | 0x (Rank: top 96%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL005467 | 1e-87 | 81.31% | |
| Bombyx | BGIBMGA011189-TA | 3e-87 | 80.81% | |
| Drosophila | Syt7-PA | 2e-78 | 73.06% | |
| EBI UniRef50 | UniRef50_UPI000237F722 | 3e-77 | 75.40% | UPI000237F722 related cluster n=1 Tax=Drosophila melanogaster RepID=UPI000237F722 |
| NCBI RefSeq | XP_392664.2 | 5e-84 | 81.91% | PREDICTED: similar to Syt7 CG2381-PG, isoform G [Apis mellifera] |
| NCBI nr blastp | gi|380028658 | 4e-83 | 81.91% | PREDICTED: synaptotagmin-7-like [Apis florea] |
| NCBI nr blastx | gi|380028658 | 1e-80 | 81.91% | PREDICTED: synaptotagmin-7-like [Apis florea] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005515 | 4.7e-26 | protein binding | |
| KEGG pathway | ||||
| InterPro domain | [18-107] IPR008973 | 4.7e-26 | C2 calcium/lipid-binding domain, CaLB | |
| [42-105] IPR000008 | 1.1e-19 | C2 calcium-dependent membrane targeting | ||
| [56-68] IPR020477 | 2.9e-07 | C2 region | ||
| Orthology group | ||||
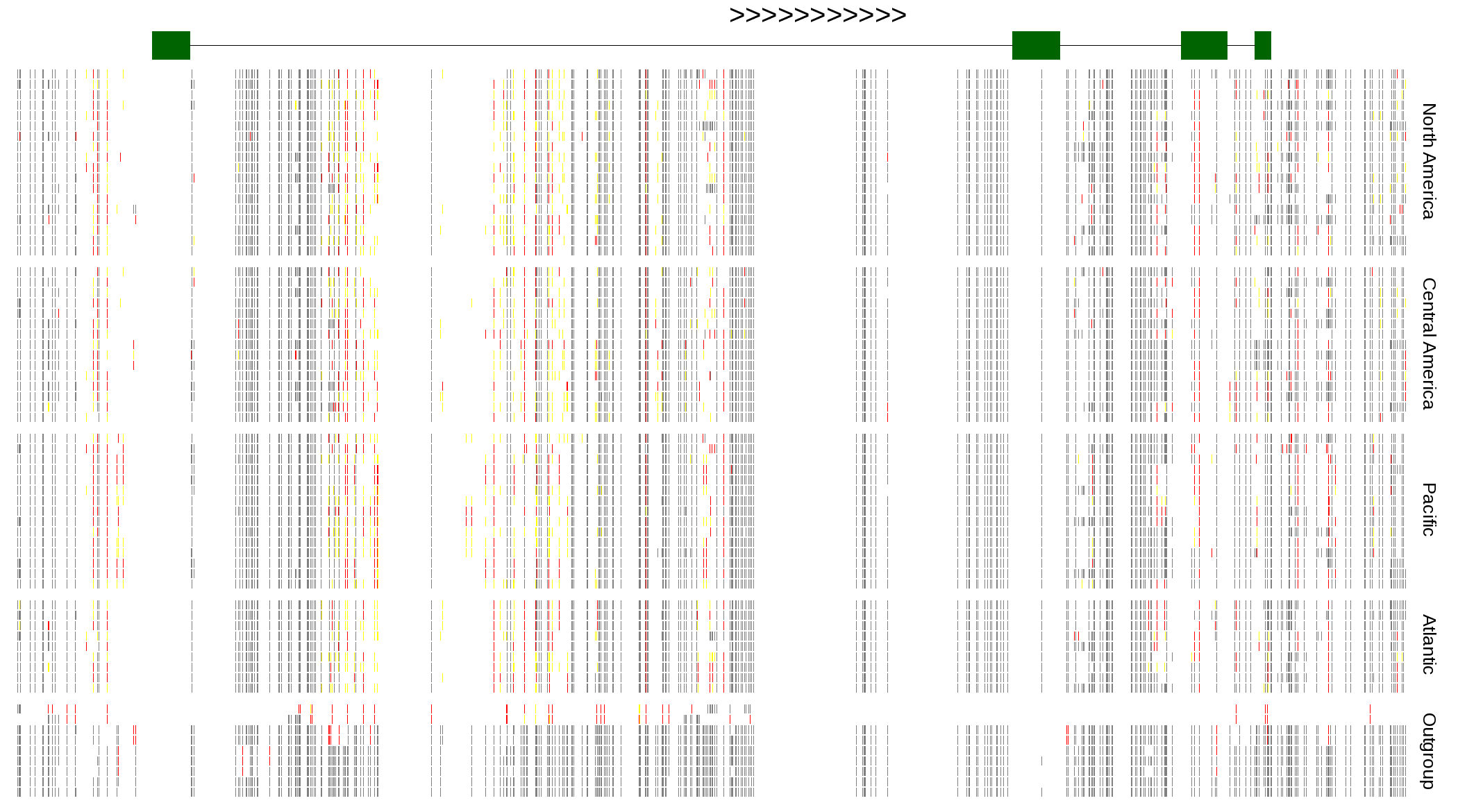
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS202972-TA
ATGGCGTTCCTACAGAACCGGTCCATCTCGCTGGTGGACATGTACATCGATAATACCGAACCGTCGGAGAACGTCGGCCAAATCCACTTCTCTTTGGAATACGACTTCCAGAATACCACTCTCATCCTCAGGATCATTCAGGGCAAGGAACTGCCGGCTAAAGATCTCAGCGGAACCTCGGACCCGTATGTCCGGGTTACGCTACTCCCGGATAAAAAACATCGCCTGGAGACCAAGATCAAACGCAGAACCCTAAATCCTCGATGGAATGAGACCTTCTATTTTGAAGGTTTCCCCATACAGAAGTTACAGAGTCGAGTGGATCTCAGCGAGAAGCCTTCTTTCTGGAAATCTCTGAAGCCCCCGGCAAAGGACAAATGTGGCGAATTGCTCACATCTCTCTGCTATCATCCCAGCAACAGCGTTCTTACCCTCACTCTGCTGAAAGCCAGGAACCTGAAGGCCAAGGACATTAACGGAAAATCGGCTGTCAACATTCATAAATTTTTAAGATCAGCTGGTATAACAACATTGACGTACGAATGA
>DPOGS202972-PA
MAFLQNRSISLVDMYIDNTEPSENVGQIHFSLEYDFQNTTLILRIIQGKELPAKDLSGTSDPYVRVTLLPDKKHRLETKIKRRTLNPRWNETFYFEGFPIQKLQSRVDLSEKPSFWKSLKPPAKDKCGELLTSLCYHPSNSVLTLTLLKARNLKAKDINGKSAVNIHKFLRSAGITTLTYE-