| DPOGS203488 | ||
|---|---|---|
| Transcript | DPOGS203488-TA | 630 bp |
| Protein | DPOGS203488-PA | 209 aa |
| Genomic position | DPSCF300055 - 841863-843989 | |
| RNAseq coverage | 460x (Rank: top 27%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL014298 | 2e-58 | 67.07% | |
| Bombyx | BGIBMGA008293-TA | 4e-61 | 61.18% | |
| Drosophila | CG1637-PB | 3e-50 | 56.29% | |
| EBI UniRef50 | UniRef50_F4W5K7 | 5e-45 | 54.66% | Iron/zinc purple acid phosphatase-like protein n=5 Tax=Coelomata RepID=F4W5K7_ACREC |
| NCBI RefSeq | XP_001651840.1 | 1e-51 | 61.11% | purple acid phosphatase, putative [Aedes aegypti] |
| NCBI nr blastp | gi|157112670 | 2e-50 | 61.11% | purple acid phosphatase, putative [Aedes aegypti] |
| NCBI nr blastx | gi|157112670 | 9e-51 | 61.11% | purple acid phosphatase, putative [Aedes aegypti] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0016787 | 1.9e-07 | hydrolase activity | |
| KEGG pathway | afm:AFUA_7G00800 | 1e-08 | ||
| K01078 (E3.1.3.2) | maps-> | Riboflavin metabolism | ||
| gamma-Hexachlorocyclohexane degradation | ||||
| InterPro domain | [48-111] IPR004843 | 1.9e-07 | Metallophosphoesterase domain | |
| Orthology group | ||||
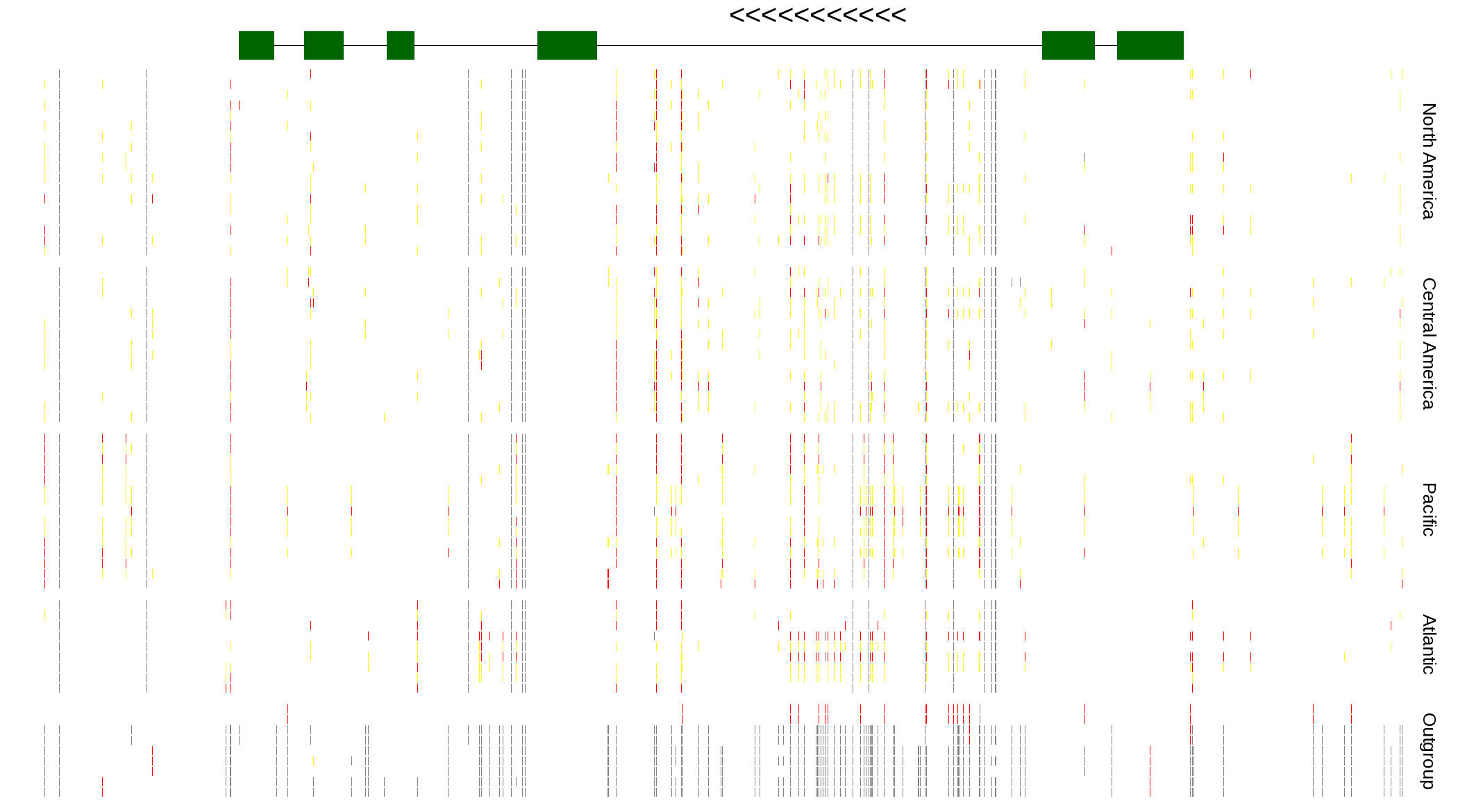
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS203488-TA
ATGCTCAGCTTCCTGTTAAAATACAATAACTGTCAGATCCCTTCGGCCGTGGGTCGTGATGCTCGGTCACCGGCCCATGTACTGCAGCGACAGGAGACTGGTCTGCTACGACCGCTGGCTCCCCAACCGCGTAGGGCTGCCCTTCCTAGGCGGGCCCGGCCTTGGCTCGTGATGTTCGGTCACCGGCCCATGTACTGCAGCAACTCGGACGACGTGGACTGCAGCGTGGAGTACACACGCAAGGGCCTGCCCTTCCTGGGCCTGTACAGTCTGGAGCCCCTGCTGAAGGAGTACCACGTGGACCTGGTGGTGTGGGCCCACGAGCACAGCTACGAGCGGTCGTGGCCGCTGTATGACGGACGAGTCTACAACGGGACGGAAGGAGCGTACGTCAACCCGCGGGCCCCGGTCCACGTGGTGACAGGTTCCGCCGGATGTCAGGAGGACACTGATAAATTCCAACGAGTCCCCCCGGAGTGGTCCGCCTTCAGGTCCAGCGACTACGGCTACACCAGACTGGCGGCCGACCGCACCGCCATACACATACAGCAGGTGGACGTGGACCTGAGGGGCCAGGTCATAGACTCCTTCACCATCGTGAAGGACCGACACGTGCCCTTTGATGTGTGA
>DPOGS203488-PA
MLSFLLKYNNCQIPSAVGRDARSPAHVLQRQETGLLRPLAPQPRRAALPRRARPWLVMFGHRPMYCSNSDDVDCSVEYTRKGLPFLGLYSLEPLLKEYHVDLVVWAHEHSYERSWPLYDGRVYNGTEGAYVNPRAPVHVVTGSAGCQEDTDKFQRVPPEWSAFRSSDYGYTRLAADRTAIHIQQVDVDLRGQVIDSFTIVKDRHVPFDV-