| DPOGS203884 | ||
|---|---|---|
| Transcript | DPOGS203884-TA | 453 bp |
| Protein | DPOGS203884-PA | 150 aa |
| Genomic position | DPSCF300402 + 83933-84894 | |
| RNAseq coverage | 805x (Rank: top 16%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL008133 | 8e-82 | 95.33% | |
| Bombyx | BGIBMGA003820-TA | 6e-80 | 92.67% | |
| Drosophila | Rpb8-PA | 1e-72 | 86.67% | |
| EBI UniRef50 | UniRef50_Q9VNZ3 | 1e-70 | 86.67% | GM03174p n=21 Tax=Metazoa RepID=Q9VNZ3_DROME |
| NCBI RefSeq | XP_970133.1 | 1e-72 | 86.67% | PREDICTED: similar to Rpb8 CG11246-PA [Tribolium castaneum] |
| NCBI nr blastp | gi|328784260 | 1e-72 | 86.75% | PREDICTED: sodium/potassium-transporting ATPase subunit alpha [Apis mellifera] |
| NCBI nr blastx | gi|383859891 | 2e-71 | 87.42% | PREDICTED: DNA-directed RNA polymerases I, II, and III subunit RPABC3-like [Megachile rotundata] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0006351 | 6.4e-98 | transcription, DNA-dependent | |
| KEGG pathway | tca:658676 | 3e-72 | ||
| K03016 (RPB8) | maps-> | Huntington's disease | ||
| Purine metabolism | ||||
| Pyrimidine metabolism | ||||
| RNA polymerase | ||||
| InterPro domain | [1-150] IPR005570 | 6.4e-98 | RNA polymerase, Rpb8 | |
| [2-147] IPR012340 | 7.3e-52 | Nucleic acid-binding, OB-fold | ||
| [2-147] IPR016027 | 1.3e-46 | Nucleic acid-binding, OB-fold-like | ||
| Orthology group | MCL13068 | Single-copy universal gene | ||
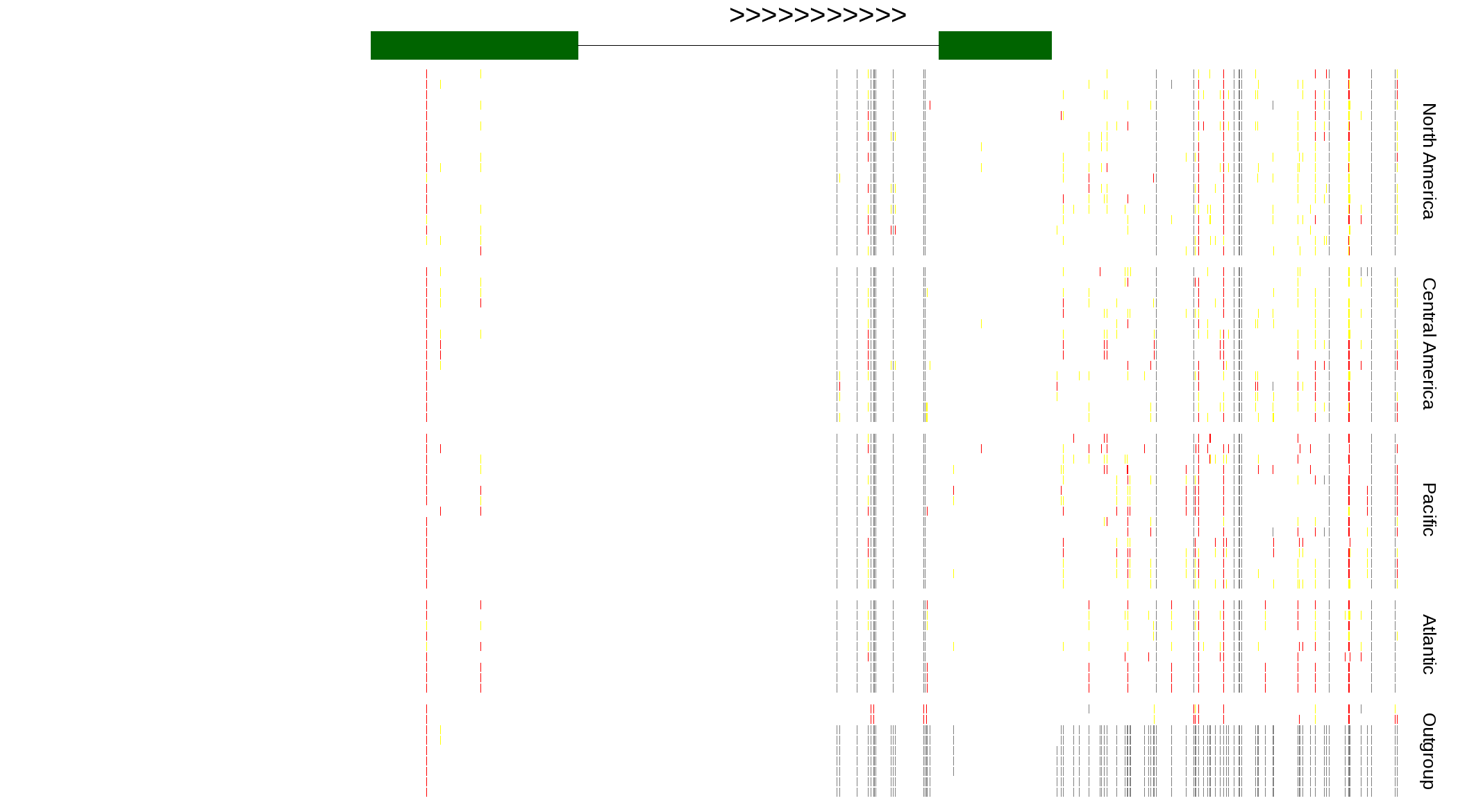
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS203884-TA
ATGGCGGGAGTGTTGTTCGAAGATATATTCAACGTCAAAGACATGGACCAGGGCGGTAAGAAATTCGATCGTGTGAGCAGATTGCATTGTGAATCAGAATCGTTTAAAATGGATCTCATTTTGGATATAAATTCTTGGATATATCCTATGGAGCTGGGAGATAAGTTCAGATTGGTTTTGGCCACAACATTACGAGAAAACGGATATCCGGATGGCGGTGAATGGAATCCTCTCGAAACAGAGGGAAATAGAGCTGATAGCTTTGAATATGTTATGTACGGAAAAGTTTATAGACTAGATGGTGATGAATTGGCCATGGAACCGACATCCCGACTGTCAGCTTTTGTTTCATTCGGTGGACTCCTCATGCGGTTGCAGGGTGACGCCAACAACCTGCACGGCTTCGAAGTCGACCAGCACATGTACTTGTTAATGAAGAAACTAGCTTTTTAA
>DPOGS203884-PA
MAGVLFEDIFNVKDMDQGGKKFDRVSRLHCESESFKMDLILDINSWIYPMELGDKFRLVLATTLRENGYPDGGEWNPLETEGNRADSFEYVMYGKVYRLDGDELAMEPTSRLSAFVSFGGLLMRLQGDANNLHGFEVDQHMYLLMKKLAF-