| DPOGS204281 | ||
|---|---|---|
| Transcript | DPOGS204281-TA | 444 bp |
| Protein | DPOGS204281-PA | 147 aa |
| Genomic position | DPSCF300046 + 90083-90526 | |
| RNAseq coverage | 285x (Rank: top 38%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL003313 | 1e-47 | 59.59% | |
| Bombyx | BGIBMGA007557-TA | 1e-44 | 57.24% | |
| Drosophila | CG4565-PB | 5e-32 | 47.52% | |
| EBI UniRef50 | UniRef50_F4X1W2 | 1e-30 | 46.85% | Histone-lysine N-methyltransferase SETMAR n=5 Tax=Formicidae RepID=F4X1W2_ACREC |
| NCBI RefSeq | XP_001604764.1 | 1e-34 | 50.35% | PREDICTED: similar to rCG56163 [Nasonia vitripennis] |
| NCBI nr blastp | gi|307177043 | 2e-31 | 47.18% | Histone-lysine N-methyltransferase SETMAR [Camponotus floridanus] |
| NCBI nr blastx | gi|340726752 | 6e-32 | 47.18% | PREDICTED: histone-lysine N-methyltransferase SETMAR-like [Bombus terrestris] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005515 | 2e-23 | protein binding | |
| KEGG pathway | dme:Dmel_CG4565 | 2e-30 | ||
| K11433 (SETMAR) | maps-> | Lysine degradation | ||
| InterPro domain | [1-121] IPR001214 | 2e-23 | SET domain | |
| Orthology group | MCL16970 | Patchy | ||
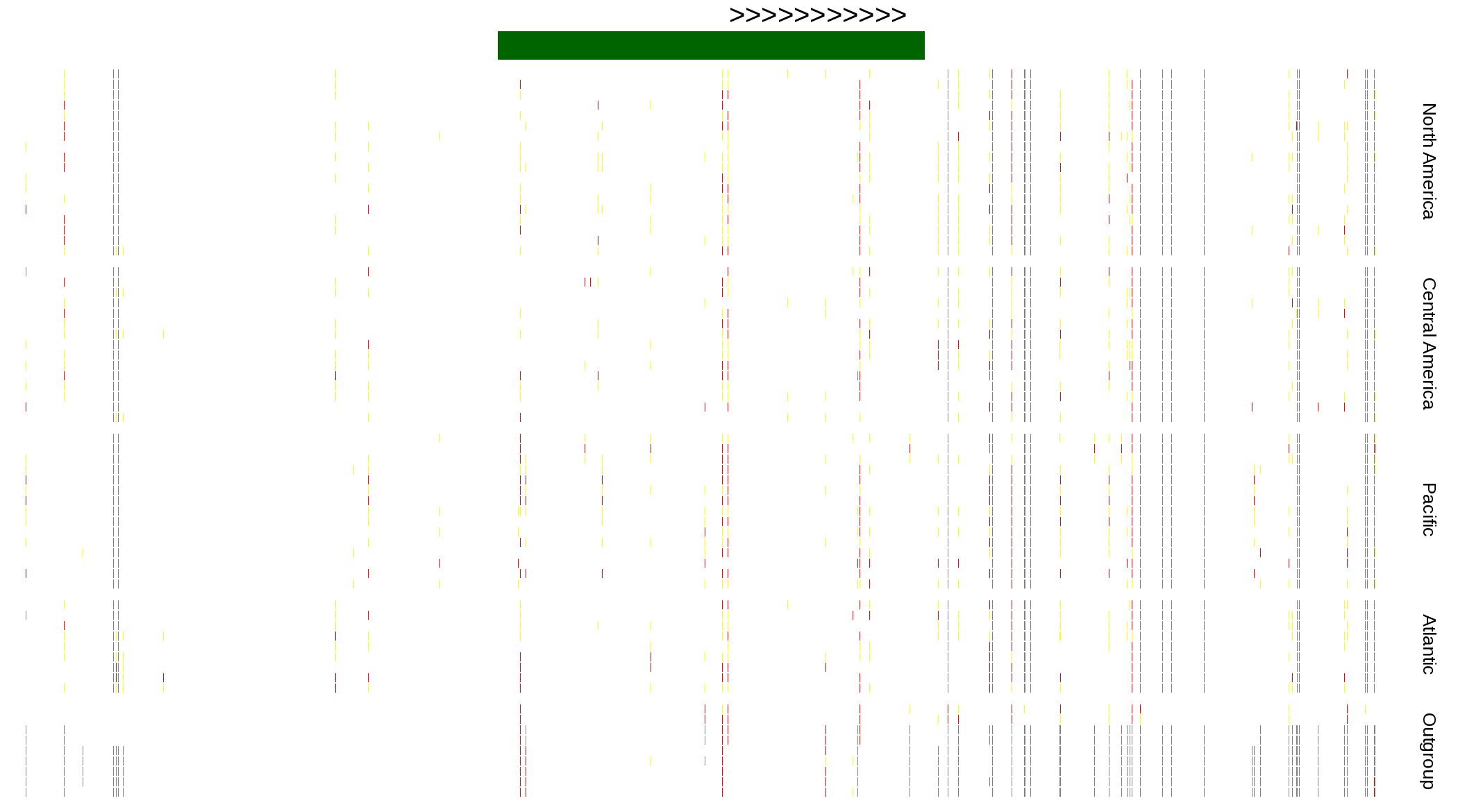
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS204281-TA
ATGGGTCTGGGTTTATTCACTGAGAAATTCATTACAGCTGGAACATTTATATGTGAATATGCTGGGGAAATCATCACTAAAACTCAGGCTCTGTACCGTCAAAAGCTAAATAAAATCCAGGGCAATATGAATTATATATTTTGTTTGAATGAATTTGTCGATGGCAGAGCTAACACAAACTTCATTGATCCTAGTCATTTTGGTAATATCGGACGTTATATTAACCACAGCTGTGACCCTAATTCAGAAATTATGCCTGTGAGAGTTGACTGTCCGATTCCTAAACTGGCTATATTCAGTTGCATTGATATATCACCGGATGAAGAAATAACATTTAATTACGGATTAAACAGTATGAGTATTACCGATGAGACCATTGAAAGAGTACCATGTCTTTGTCAGAGTGCTAAATGTAAAGGCTTTATACCGTTAGATAGTTATTGA
>DPOGS204281-PA
MGLGLFTEKFITAGTFICEYAGEIITKTQALYRQKLNKIQGNMNYIFCLNEFVDGRANTNFIDPSHFGNIGRYINHSCDPNSEIMPVRVDCPIPKLAIFSCIDISPDEEITFNYGLNSMSITDETIERVPCLCQSAKCKGFIPLDSY-