| DPOGS204314 | ||
|---|---|---|
| Transcript | DPOGS204314-TA | 507 bp |
| Protein | DPOGS204314-PA | 168 aa |
| Genomic position | DPSCF300046 + 710876-714865 | |
| RNAseq coverage | 543x (Rank: top 23%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL015144 | 2e-59 | 67.26% | |
| Bombyx | BGIBMGA007505-TA | 5e-53 | 85.19% | |
| Drosophila | RhoL-PB | 4e-60 | 60.00% | |
| EBI UniRef50 | UniRef50_F5HKZ0 | 2e-59 | 63.31% | AGAP003092-PA n=2 Tax=Anopheles RepID=F5HKZ0_ANOGA |
| NCBI RefSeq | XP_001238816.1 | 4e-60 | 63.31% | AGAP003092-PA [Anopheles gambiae str. PEST] |
| NCBI nr blastp | gi|118783126 | 6e-59 | 63.31% | AGAP003092-PA [Anopheles gambiae str. PEST] |
| NCBI nr blastx | gi|328791872 | 3e-57 | 65.87% | PREDICTED: ras-like GTP-binding protein RhoL-like [Apis mellifera] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0007264 | 9.5e-90 | small GTPase mediated signal transduction | |
| GO:0005622 | 9.5e-90 | intracellular | ||
| GO:0005525 | 9.5e-90 | GTP binding | ||
| GO:0015031 | 3e-23 | protein transport | ||
| GO:0016020 | 9.1e-16 | membrane | ||
| GO:0007165 | 9.1e-16 | signal transduction | ||
| GO:0006184 | 9.1e-16 | GTP catabolic process | ||
| GO:0003924 | 9.1e-16 | GTPase activity | ||
| KEGG pathway | tgu:100223508 | 9e-51 | ||
| K07863 (RHOG) | maps-> | Shigellosis | ||
| Bacterial invasion of epithelial cells | ||||
| InterPro domain | [1-157] IPR003578 | 9.5e-90 | Small GTPase superfamily, Rho type | |
| [2-155] IPR001806 | 2.1e-50 | Small GTPase superfamily | ||
| [2-149] IPR005225 | 7.6e-31 | Small GTP-binding protein domain | ||
| [2-157] IPR003579 | 3e-23 | Small GTPase superfamily, Rab type | ||
| [2-157] IPR020849 | 9.1e-16 | Small GTPase superfamily, Ras type | ||
| Orthology group | MCL18324 | Insect specific | ||
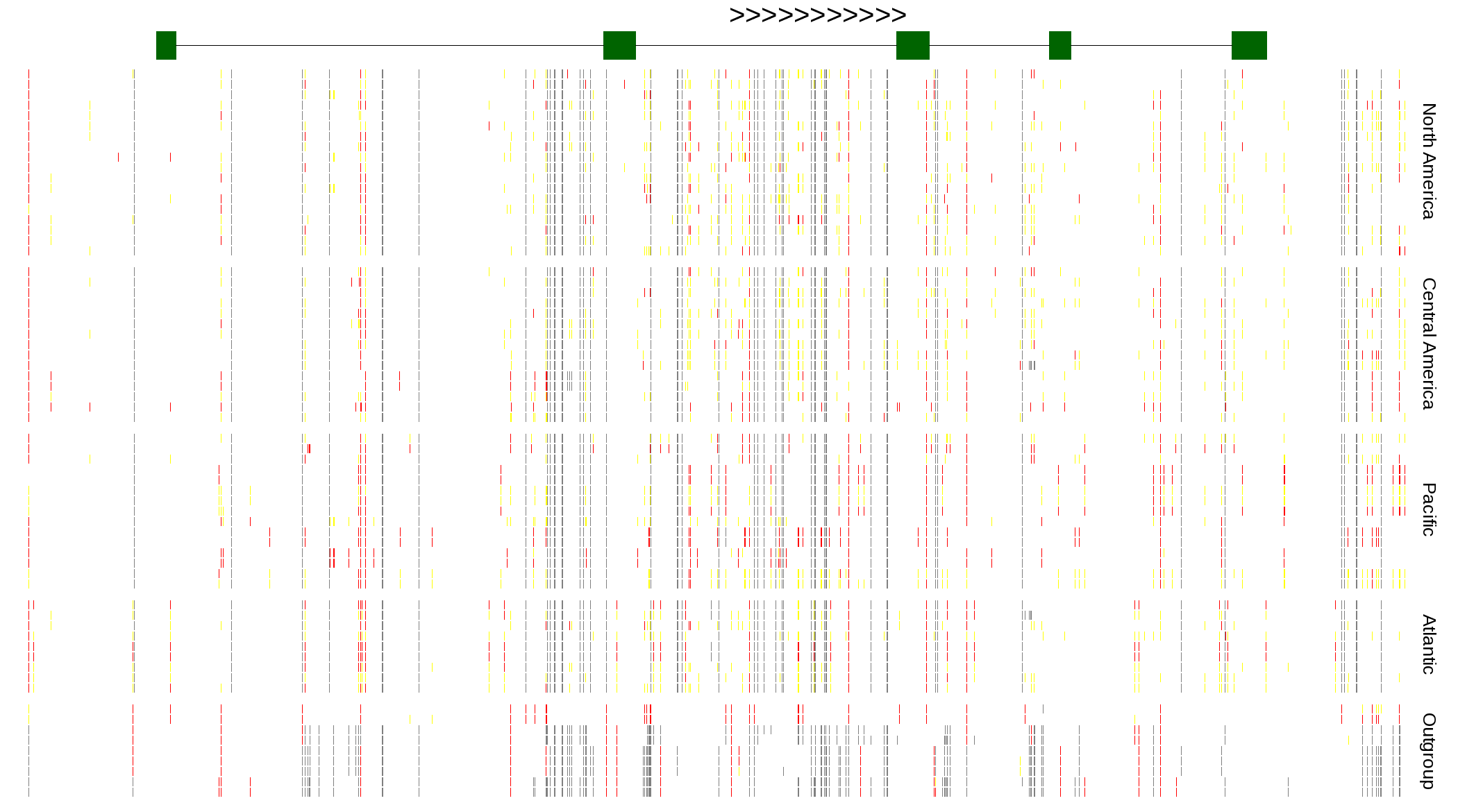
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS204314-TA
ATGGTTGGCAAAACTTGCCTTCTCTACGTTTACACTAGAAACGAATTCCCTAAAGAATACGTACCAACGGTATTTGACAATTATGTAGGACACATTACGGTTGATGGACAAGAAATTGAAATGACACTCTGGGACACTGCCGGCCAGGAAGATTACGAAAGACTTAGACCTTTGTCATATAATAACACGCATTGTTTCCTGGTATGTTACTCAGTTGGGAGTAGATCGTCTTATGAAAACGTTGGTCACAAATGGTATCCTGAATTAAAACATTTCAGTTCGTCAGTGCCAATAGTTCTTGTTGCTACTAAAACAGATTTAAGGGCAAGCGGTCAAGCGGTTATTACAACTCAAGAAGGAAAAGTATTAAAAAGGAAAATAAGAGCAGTCCAATTGGTTGAATGTTCTGCTTTGGAACGAGAGAATATGCATGAAGTATTCGAGGAGGCCGTGCGTGCTGCTCTCAAGAAGAAGCAAGTAACAAAACGAACTTGCAATTTCCTATGA
>DPOGS204314-PA
MVGKTCLLYVYTRNEFPKEYVPTVFDNYVGHITVDGQEIEMTLWDTAGQEDYERLRPLSYNNTHCFLVCYSVGSRSSYENVGHKWYPELKHFSSSVPIVLVATKTDLRASGQAVITTQEGKVLKRKIRAVQLVECSALERENMHEVFEEAVRAALKKKQVTKRTCNFL-