| DPOGS205226 | ||
|---|---|---|
| Transcript | DPOGS205226-TA | 255 bp |
| Protein | DPOGS205226-PA | 84 aa |
| Genomic position | DPSCF300265 + 106841-110914 | |
| RNAseq coverage | 9923x (Rank: top 1%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL006471 | 4e-38 | 94.87% | |
| Bombyx | BGIBMGA014507-TA | 8e-17 | 73.47% | |
| Drosophila | VhaM9.7-a-PB | 2e-24 | 56.79% | |
| EBI UniRef50 | UniRef50_Q0IEF0 | 3e-27 | 69.14% | Vacuolar ATP synthase subunit H n=24 Tax=Pancrustacea RepID=Q0IEF0_AEDAE |
| NCBI RefSeq | XP_624787.1 | 2e-28 | 71.25% | PREDICTED: similar to CG1268-PA [Apis mellifera] |
| NCBI nr blastp | gi|3687299 | 1e-36 | 86.90% | vacuolar ATPase, subunit M9.7 [Manduca sexta] |
| NCBI nr blastx | gi|3687299 | 2e-40 | 90.12% | vacuolar ATPase, subunit M9.7 [Manduca sexta] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0033179 | 8.8e-55 | proton-transporting V-type ATPase, V0 domain | |
| GO:0015991 | 8.8e-55 | ATP hydrolysis coupled proton transport | ||
| GO:0015078 | 8.8e-55 | hydrogen ion transmembrane transporter activity | ||
| KEGG pathway | aga:AgaP_AGAP003588 | 2e-27 | ||
| K02153 (ATPeVH, ATP6H) | maps-> | Collecting duct acid secretion | ||
| Oxidative phosphorylation | ||||
| Phagosome | ||||
| Vibrio cholerae infection | ||||
| Epithelial cell signaling in Helicobacter pylori infection | ||||
| InterPro domain | [1-84] IPR017385 | 8.8e-55 | ATPase, V0 complex, subunit E, metazoa | |
| [1-83] IPR008389 | 1.4e-39 | ATPase, V0 complex, subunit E | ||
| Orthology group | MCL12381 | Multiple-copy universal gene | ||
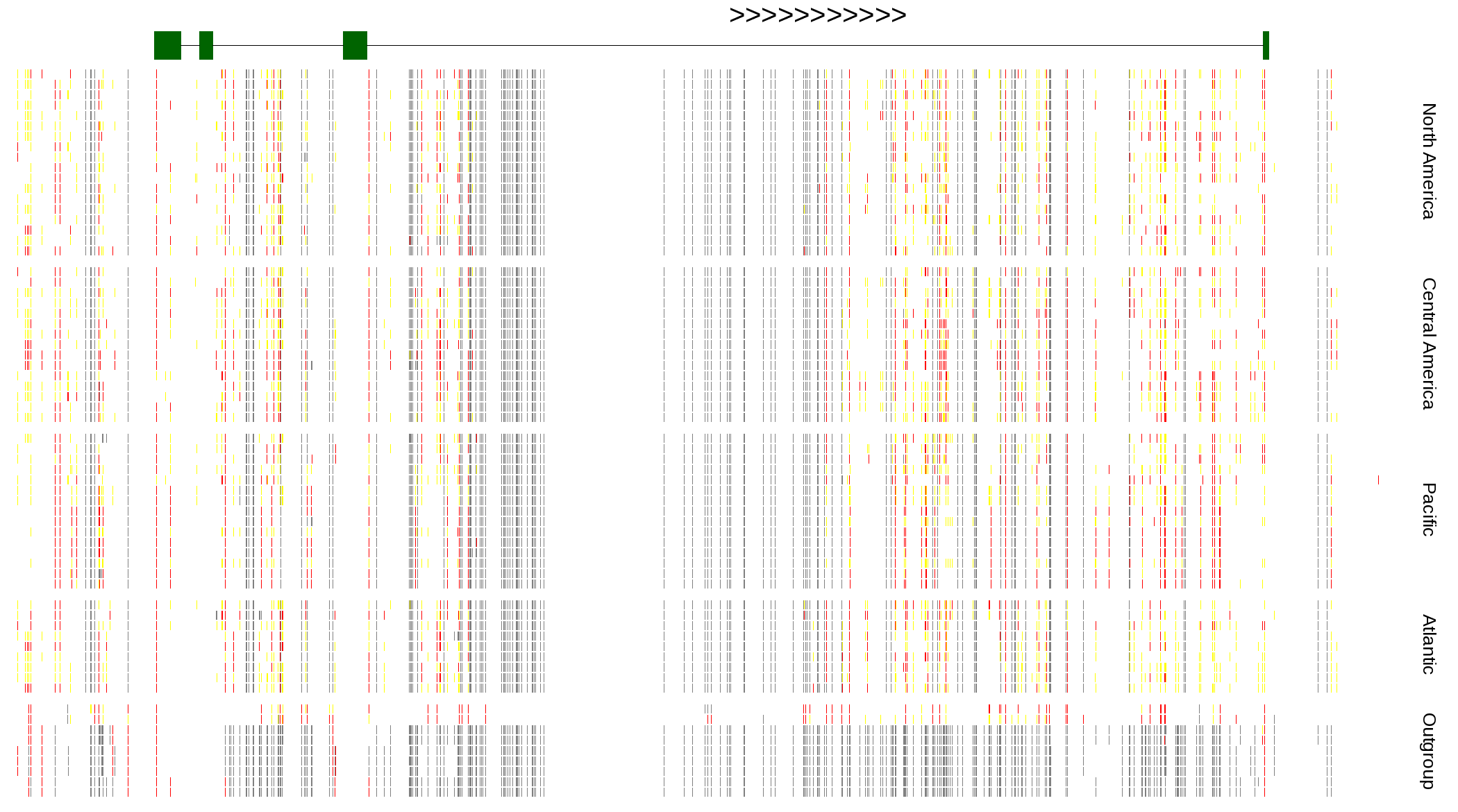
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS205226-TA
ATGGGTGCTTCATTCGTTCCCATCACCATTTTCACCGTCTTCTGGGGGGTCATTGGTATTGTGTGCCCCTTCTTCGCGCCCAAAGGACCTAACCGAGGGATTATCCAAGTGGTGTTGATGTTAACAGCGGCTACGTGCTGGTTGTTCTGGTTGTGTGCGTACATGGCACAAATGAATCCTCTCGTCGGACCGAGACTCAGCAACGAAACTCTCATCTGGATAGCTCGCACATGGGGTAATCCATACAACAAGTAA
>DPOGS205226-PA
MGASFVPITIFTVFWGVIGIVCPFFAPKGPNRGIIQVVLMLTAATCWLFWLCAYMAQMNPLVGPRLSNETLIWIARTWGNPYNK-