| DPOGS205381 | ||
|---|---|---|
| Transcript | DPOGS205381-TA | 651 bp |
| Protein | DPOGS205381-PA | 216 aa |
| Genomic position | DPSCF300373 + 80246-84698 | |
| RNAseq coverage | 20068x (Rank: top 0%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL013442 | 6e-109 | 84.72% | |
| Bombyx | BGIBMGA008768-TA | 4e-71 | 70.00% | |
| Drosophila | Fer1HCH-PG | 1e-54 | 57.14% | |
| EBI UniRef50 | UniRef50_A8D916 | 2e-60 | 53.21% | Ferritin n=3 Tax=Endopterygota RepID=A8D916_BOMIG |
| NCBI RefSeq | NP_001037580.1 | 1e-79 | 64.81% | ferritin [Bombyx mori] |
| NCBI nr blastp | gi|124245059 | 4e-97 | 78.64% | ferritin HCH [Pieris rapae] |
| NCBI nr blastx | gi|5738076 | 2e-95 | 78.24% | secreted ferritin S subunit precursor [Calpodes ethlius] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0006879 | 6e-74 | cellular iron ion homeostasis | |
| GO:0006826 | 6e-74 | iron ion transport | ||
| GO:0008199 | 6e-74 | ferric iron binding | ||
| GO:0005488 | 6e-74 | binding | ||
| GO:0046914 | 2.1e-55 | transition metal ion binding | ||
| GO:0055114 | 2.1e-55 | oxidation-reduction process | ||
| GO:0016491 | 2.1e-55 | oxidoreductase activity | ||
| KEGG pathway | tca:655420 | 2e-61 | ||
| K00522 (FTH1) | maps-> | Porphyrin and chlorophyll metabolism | ||
| InterPro domain | [41-216] IPR001519 | 6e-74 | Ferritin | |
| [23-213] IPR012347 | 1e-57 | Ferritin-related | ||
| [17-215] IPR009078 | 2.1e-55 | Ferritin/ribonucleotide reductase-like | ||
| [46-195] IPR008331 | 9.7e-44 | Ferritin/DPS protein domain | ||
| Orthology group | MCL15901 | Insect specific | ||
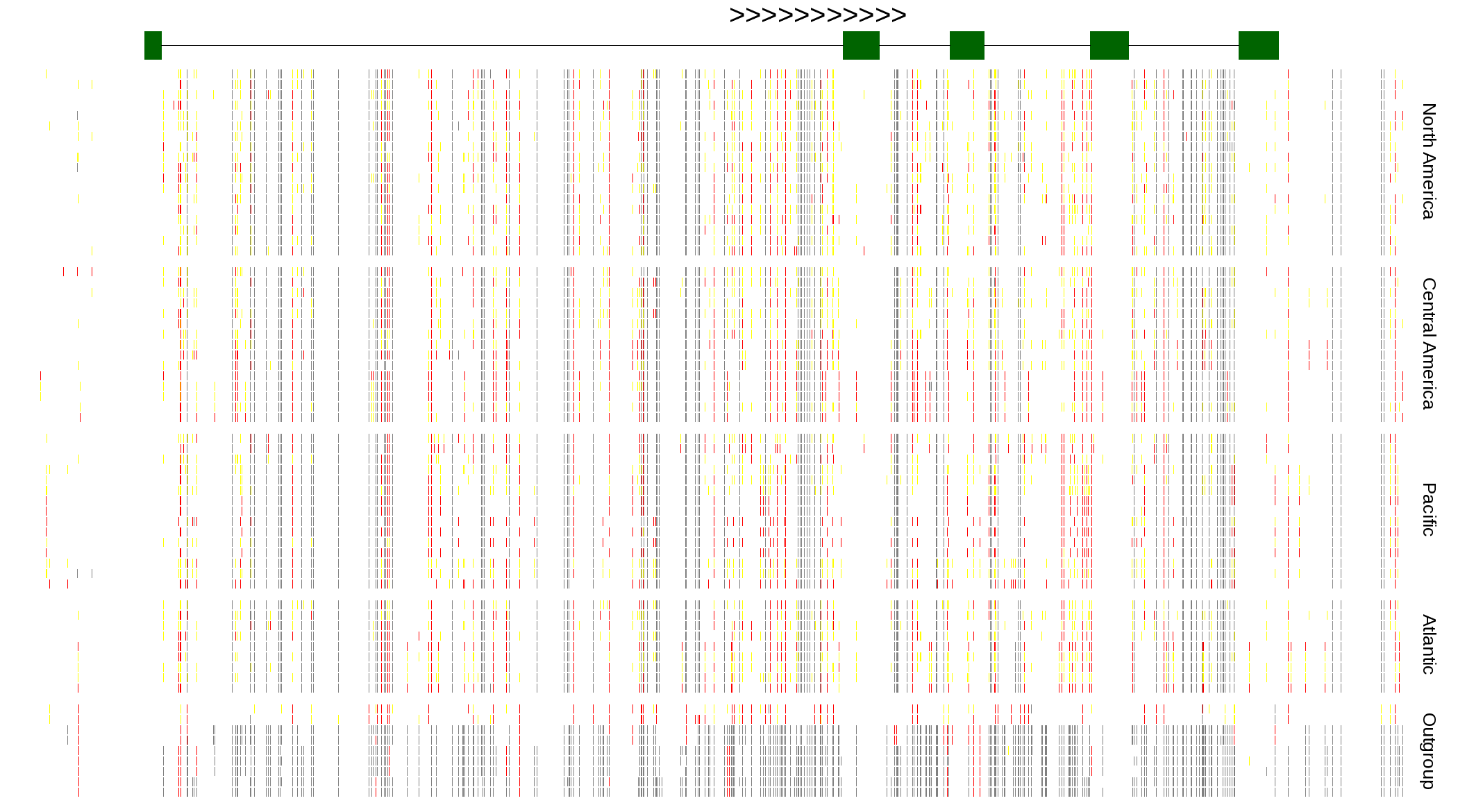
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS205381-TA
ATGAAGACCTTCTTGTTTGTCATCCTCGCTGTCCTGGCTGTTTGCCCTTCGGCTTTCGGCACACAATGTAATGTCAACCCGGTCAATATACCTAAGAATTGGATCACCATGACCCACGGCTGCCGCGACAGTGTCCGCCGTCAGATACAGCTGGAGGTGGCGGCTTCAGTGCAGTACCTCGCGATGGGAGCCTACTTCTCCAAGGATAGCGTGAACAGGCCAGGTTTCGCGAAGCTGTTCTTCGACGCCAGTTCTGAGGAACGCGAGCACGCTATGAAGCTGATCGAATACCTTCTGATGAGGGGAGAACTAACAAACAGTGTGTCCTCGCTTATCACTATTAGGCCCCCAGAACACAAGCACTGGGTCAGCGGACAGGAAGCCCTGGAACATGCGCTGAAGTTGGAGACGGAAGTCACAAAGGCCATTAAGAATGTTATCATCAAGTGTGAGAACGACAGAGACGGCACCGGAGAAGGCAACAACGATTACCATCTCGTTGATTACTTGACCGGCGAGTTCCTTGAGGAGCAGTACAAGGGACAGAGAGACCTCGCCGGCAAGGCCAGCACCCTCAAGAAGATGATGGACCGTCACTCAGCCCTCGGAGAGTTCATCTTTGACAAGAAACTCCTCGGAATTGACATTTAA
>DPOGS205381-PA
MKTFLFVILAVLAVCPSAFGTQCNVNPVNIPKNWITMTHGCRDSVRRQIQLEVAASVQYLAMGAYFSKDSVNRPGFAKLFFDASSEEREHAMKLIEYLLMRGELTNSVSSLITIRPPEHKHWVSGQEALEHALKLETEVTKAIKNVIIKCENDRDGTGEGNNDYHLVDYLTGEFLEEQYKGQRDLAGKASTLKKMMDRHSALGEFIFDKKLLGIDI-