| DPOGS206439 | ||
|---|---|---|
| Transcript | DPOGS206439-TA | 522 bp |
| Protein | DPOGS206439-PA | 173 aa |
| Genomic position | DPSCF300070 - 731627-732148 | |
| RNAseq coverage | 363x (Rank: top 33%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL012713 | 5e-92 | 89.60% | |
| Bombyx | BGIBMGA005444-TA | 4e-63 | 84.38% | |
| Drosophila | CG9547-PA | 1e-79 | 77.33% | |
| EBI UniRef50 | UniRef50_Q92947 | 1e-76 | 76.74% | Glutaryl-CoA dehydrogenase, mitochondrial n=544 Tax=root RepID=GCDH_HUMAN |
| NCBI RefSeq | XP_001663829.1 | 6e-84 | 83.72% | acyl-coa dehydrogenase [Aedes aegypti] |
| NCBI nr blastp | gi|157136755 | 8e-83 | 83.72% | acyl-coa dehydrogenase [Aedes aegypti] |
| NCBI nr blastx | gi|157136755 | 2e-79 | 83.72% | acyl-coa dehydrogenase [Aedes aegypti] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0016627 | 6.7e-44 | oxidoreductase activity, acting on the CH-CH group of donors | |
| GO:0055114 | 6.7e-44 | oxidation-reduction process | ||
| KEGG pathway | aag:AaeL_AAEL013642 | 2e-83 | ||
| K00252 (E1.3.99.7, gcdH) | maps-> | Fatty acid metabolism | ||
| Benzoate degradation via CoA ligation | ||||
| Tryptophan metabolism | ||||
| Lysine degradation | ||||
| InterPro domain | [20-170] IPR009075 | 6.7e-44 | Acyl-CoA dehydrogenase/oxidase C-terminal | |
| [26-166] IPR006090 | 4.6e-29 | Acyl-CoA oxidase/dehydrogenase, type 1 | ||
| Orthology group | MCL11749 | Single-copy universal gene | ||
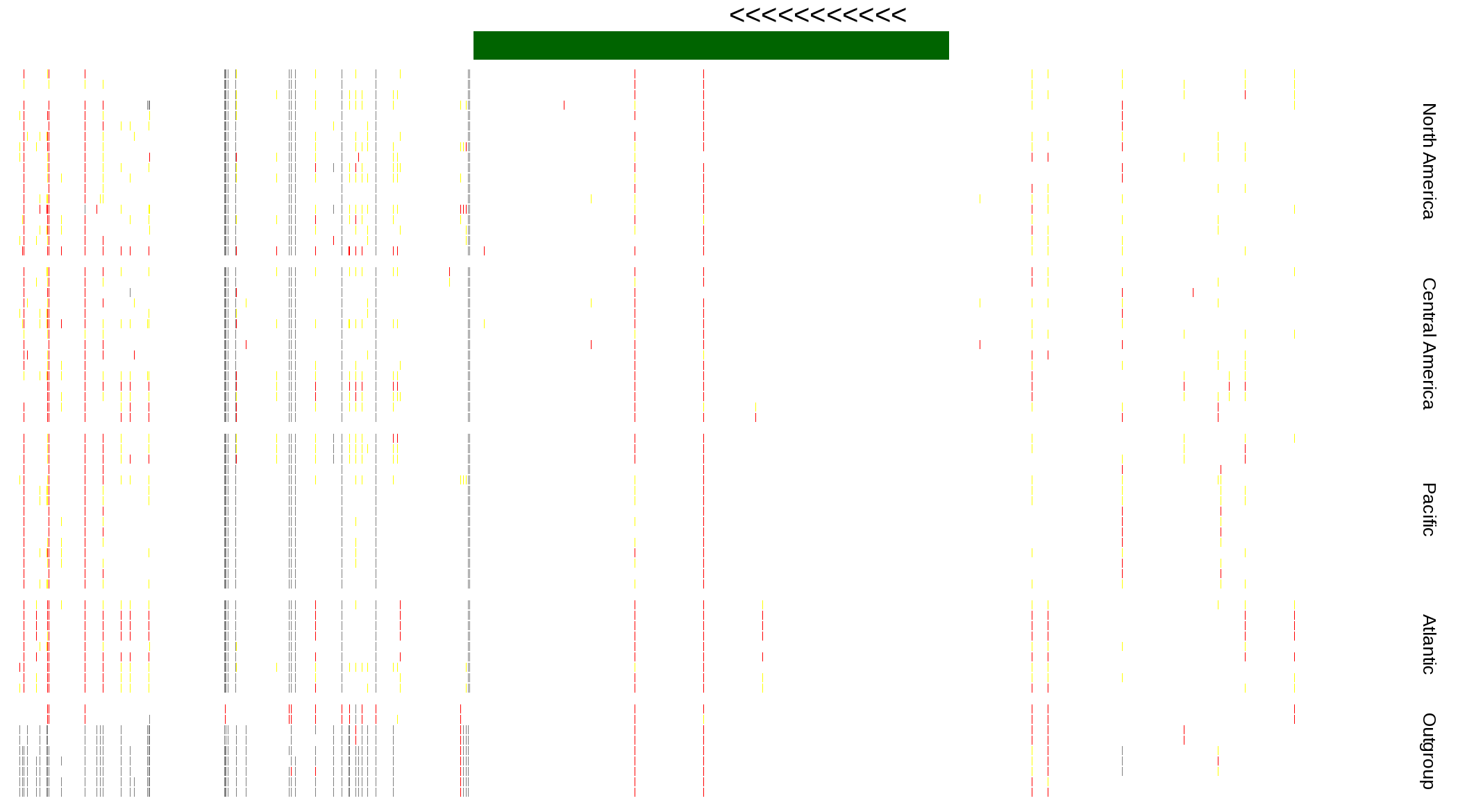
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS206439-TA
ATGATATTGCTCGACGAAGTAGTTATACCAGAGGAGAGTTTATTACCCGGGGTATTGGGTATGAAGGGGCCTTTCGGATGTCTCAATAATGCGAGATACGGGATCGCTTGGGGCGCTCTCGGTGCGGCCGAATCGTGTTTGCGGATTGCTCGACAGTACACATTAGATCGGAAACAATTTGGAAAACCGCTCGCTTCGAACCAGCTGATACAAAAGAAAATGGCGGACATGATAACTGAAATAAGTCTCGGTTTTCAAGGTTGCTTGCAAGTCGGTCGTCTAAAAGATGAGGGTTTAGTAGCTCCCGAAATGATTTCTCTTTTGAAAAGAAACAACTGCGGCAAGGCTTTAGAGATTGCGAGAGTAGCCAGGGACATGCTCGGTGGTAATGGAGTGTCTGATGAGTATCATATAATAAGACACGTTATGAATCTTGAGGCCGTTAATACTTATGAAGGGACTCATGATATCCACGCCCTAATTTTAGGAAAAGCGATAACAGGAATACAGTCTTTCTATTAA
>DPOGS206439-PA
MILLDEVVIPEESLLPGVLGMKGPFGCLNNARYGIAWGALGAAESCLRIARQYTLDRKQFGKPLASNQLIQKKMADMITEISLGFQGCLQVGRLKDEGLVAPEMISLLKRNNCGKALEIARVARDMLGGNGVSDEYHIIRHVMNLEAVNTYEGTHDIHALILGKAITGIQSFY-