| DPOGS206871 | ||
|---|---|---|
| Transcript | DPOGS206871-TA | 690 bp |
| Protein | DPOGS206871-PA | 229 aa |
| Genomic position | DPSCF300001 - 2314363-2317184 | |
| RNAseq coverage | 603x (Rank: top 21%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL003593 | 2e-55 | 78.63% | |
| Bombyx | BGIBMGA010918-TA | 6e-51 | 42.42% | |
| Drosophila | CG5326-PA | 3e-49 | 44.76% | |
| EBI UniRef50 | UniRef50_Q5TWT4 | 8e-57 | 50.22% | AGAP007264-PA n=6 Tax=Culicidae RepID=Q5TWT4_ANOGA |
| NCBI RefSeq | XP_564957.3 | 2e-57 | 50.22% | AGAP007264-PA [Anopheles gambiae str. PEST] |
| NCBI nr blastp | gi|158285959 | 3e-56 | 50.22% | AGAP007264-PA [Anopheles gambiae str. PEST] |
| NCBI nr blastx | gi|158285959 | 4e-61 | 48.90% | AGAP007264-PA [Anopheles gambiae str. PEST] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0016021 | 3e-73 | integral to membrane | |
| KEGG pathway | dme:Dmel_CG6660 | 9e-43 | ||
| K00680 (E2.3.1.-) | maps-> | Benzoate degradation via CoA ligation | ||
| Limonene and pinene degradation | ||||
| Ethylbenzene degradation | ||||
| Tyrosine metabolism | ||||
| 1- and 2-Methylnaphthalene degradation | ||||
| InterPro domain | [10-229] IPR002076 | 3e-73 | GNS1/SUR4 membrane protein | |
| Orthology group | MCL34684 | Lepidoptera specific | ||
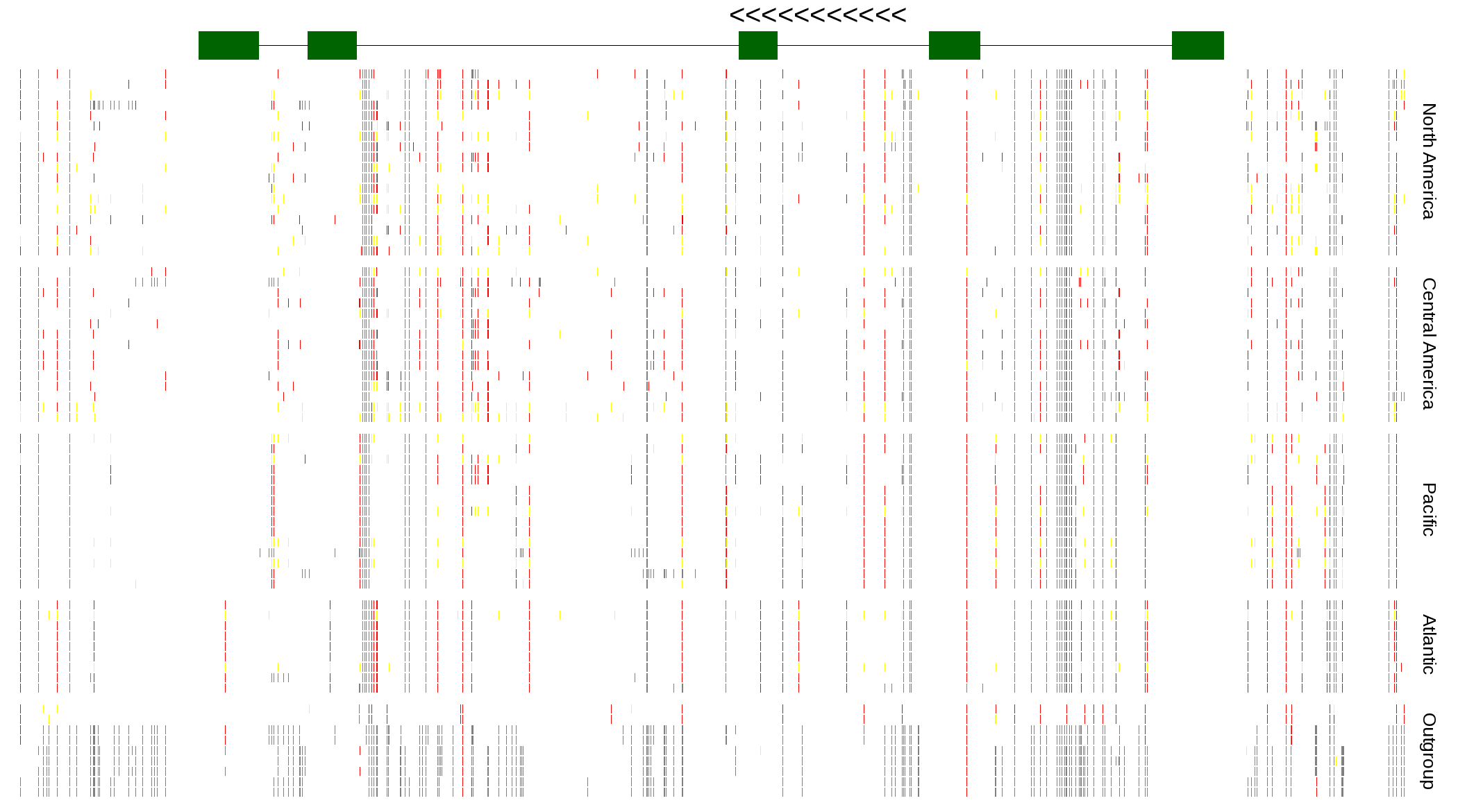
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS206871-TA
ATGTCGATCATGTTTCTATACCATCGATTTGTTAAATCTTGGGGACCAGCGTTTATGGCCAATCGACCTGCATACGATTTGAAGAGACTTCTTATTTTATACAACATCATACAGATACTCCTTTCTGGATTACTCGCATTTGAGTGTCTTCGATGGCTTTACCTACCAGGTTACTACAGTGTCTGGTGCCAGAAAATCAACTACGAAGACACGGAAGTGGAACGTATGGTCATCAATTACGTGTGGTTATATTACGCCATCAAACTTGTAGATTTATTAGATACGGTTTTCTTCGTCTTCCGTAAGAAATTCAATCAGGTATCTTTTCTGCATGTTTACCATCACCTGGGCATGTGCTTGCTGGGATACGTTGGAACTAAATACGTCCCCGGTGGACACGGTATAATGTTGGGTTTTATAAACTCCTTCGTGCACGCAGTGATGTACACCTACTATTTAATATCGGCCGTTAAACGTGAGTGGGTTAGAATATGGTGGAAGAAATACATCACACAACTACAGATTTTACAATTCCTTCTCCTGATTATACACTTCGGCCACATCTTATTTGAACCCTCATGCGGTTATCCAAAATGGGTGTCCTTTGTATTTTTACCCCACAATATATTCATATTGTTCCTATTTTCAGATTTTTACCTTAAGGAATACATTCACAAAAAGAAGAAATAA
>DPOGS206871-PA
MSIMFLYHRFVKSWGPAFMANRPAYDLKRLLILYNIIQILLSGLLAFECLRWLYLPGYYSVWCQKINYEDTEVERMVINYVWLYYAIKLVDLLDTVFFVFRKKFNQVSFLHVYHHLGMCLLGYVGTKYVPGGHGIMLGFINSFVHAVMYTYYLISAVKREWVRIWWKKYITQLQILQFLLLIIHFGHILFEPSCGYPKWVSFVFLPHNIFILFLFSDFYLKEYIHKKKK-