| DPOGS206997 | ||
|---|---|---|
| Transcript | DPOGS206997-TA | 399 bp |
| Protein | DPOGS206997-PA | 132 aa |
| Genomic position | DPSCF300001 + 896297-902684 | |
| RNAseq coverage | 19489x (Rank: top 1%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL002112 | 3e-71 | 98.48% | |
| Bombyx | BGIBMGA013111-TA | 8e-35 | 76.19% | |
| Drosophila | fabp-PC | 7e-24 | 53.39% | |
| EBI UniRef50 | UniRef50_B6CMG0 | 7e-66 | 91.67% | Fatty acid-binding protein 3 n=7 Tax=Neoptera RepID=B6CMG0_HELAM |
| NCBI RefSeq | NP_001011630.1 | 8e-37 | 56.49% | fatty acid binding protein [Apis mellifera] |
| NCBI nr blastp | gi|3115357 | 6e-67 | 94.70% | cellular retinoic acid binding protein [Manduca sexta] |
| NCBI nr blastx | gi|3115357 | 1e-65 | 94.70% | cellular retinoic acid binding protein [Manduca sexta] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005488 | 3.5e-50 | binding | |
| GO:0008289 | 1.1e-17 | lipid binding | ||
| GO:0006810 | 1.1e-17 | transport | ||
| GO:0005215 | 1.1e-17 | transporter activity | ||
| KEGG pathway | dre:171478 | 5e-25 | ||
| K08752 (FABP3) | maps-> | PPAR signaling pathway | ||
| InterPro domain | [1-130] IPR012674 | 3.5e-50 | Calycin | |
| [1-130] IPR011038 | 4.7e-45 | Calycin-like | ||
| [7-130] IPR000566 | 1.4e-19 | Lipocalin/cytosolic fatty-acid binding protein domain | ||
| [4-26] IPR000463 | 1.1e-17 | Cytosolic fatty-acid binding | ||
| Orthology group | MCL10616 | Multiple-copy universal gene | ||
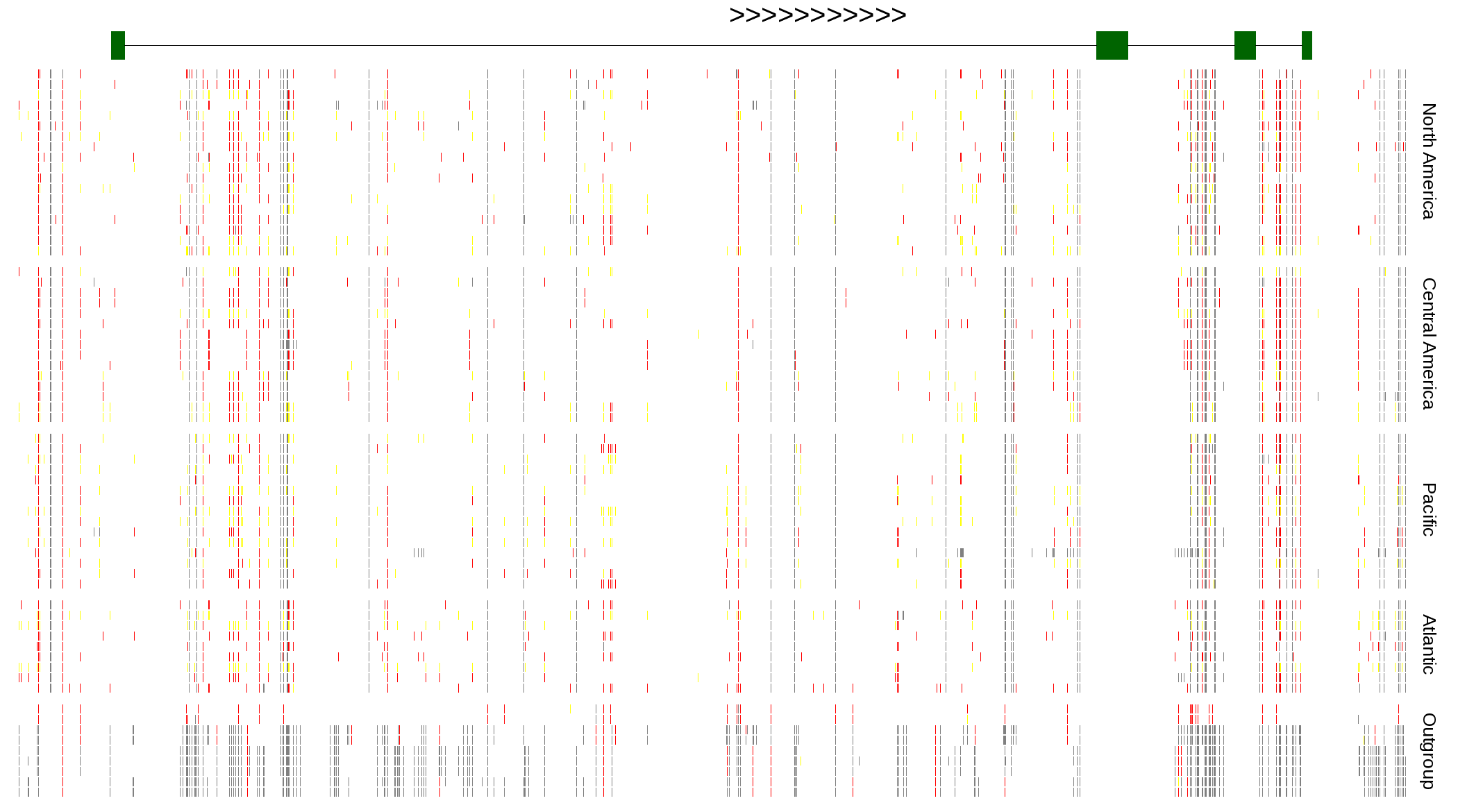
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS206997-TA
ATGGAATTCGTTGGCAAGAAATACAAAATGGTCTCCTCTGAGAACTTCGATGAGTTCATGAAGACTATTGGCGTTGGTCTCATCACCCGTAAGGCCGCTAATGCGGTAACGCCGACTGTGGAGCTGCGTCAGGACGGGGACAACTTTGTCCTAGTGACGTCATCCACCTTCAAGACCACGGAGATGAAGTTCAAGCCCGGTGAGGAATTTGACGAGGAGCGCGCTGATGGTGCCAAGGTGAAGTCCGTTTGCACCTTCGAAGGGAACACCTTGAAGCAGGTTCAGAAAGCCGCTGACGGGCTGGAGGTCACCTACGTCAGGGAATTCGGCCCCGAAGAGATGAAGGCTGTTATGACAGCTAAGGATGTAACTTGCACAAGAGTGTACAAGGTACAATAA
>DPOGS206997-PA
MEFVGKKYKMVSSENFDEFMKTIGVGLITRKAANAVTPTVELRQDGDNFVLVTSSTFKTTEMKFKPGEEFDEERADGAKVKSVCTFEGNTLKQVQKAADGLEVTYVREFGPEEMKAVMTAKDVTCTRVYKVQ-