| DPOGS207013 | ||
|---|---|---|
| Transcript | DPOGS207013-TA | 648 bp |
| Protein | DPOGS207013-PA | 215 aa |
| Genomic position | DPSCF300001 + 1165312-1169489 | |
| RNAseq coverage | 34x (Rank: top 74%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL002124 | 5e-25 | 42.34% | |
| Bombyx | BGIBMGA012942-TA | 3e-28 | 44.12% | |
| Drosophila | fabp-PC | 3e-09 | 34.78% | |
| EBI UniRef50 | UniRef50_B6CMG0 | 3e-19 | 39.10% | Fatty acid-binding protein 3 n=7 Tax=Neoptera RepID=B6CMG0_HELAM |
| NCBI RefSeq | XP_001608047.1 | 1e-15 | 37.50% | PREDICTED: similar to fatty acid binding protein isoform 1 [Nasonia vitripennis] |
| NCBI nr blastp | gi|171740911 | 9e-19 | 39.10% | fatty acid-binding protein 3 [Helicoverpa armigera] |
| NCBI nr blastx | gi|171740911 | 1e-18 | 39.39% | fatty acid-binding protein 3 [Helicoverpa armigera] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005488 | 7e-25 | binding | |
| KEGG pathway | mmu:14077 | 4e-10 | ||
| K08752 (FABP3) | maps-> | PPAR signaling pathway | ||
| InterPro domain | [2-135] IPR011038 | 2.9e-26 | Calycin-like | |
| [1-134] IPR012674 | 7e-25 | Calycin | ||
| Orthology group | ||||
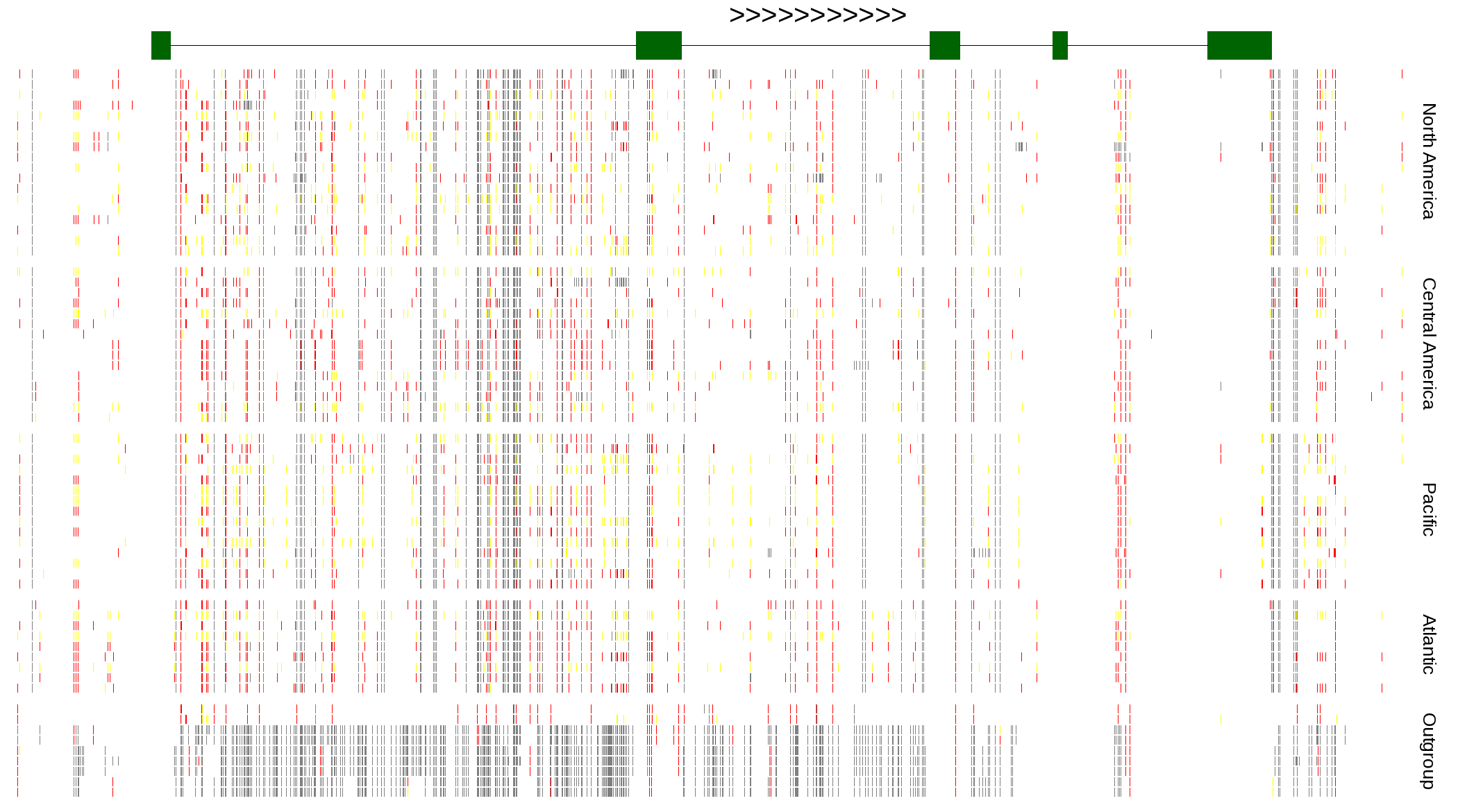
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS207013-TA
ATGGACAATTTCCTGGGCAAGGAATACGTCATGGTGTCGCAGGAGAACTTTGACGACTATCTCGTGTTTATGGGTGTGAGTTACCTTAACAGAAAACTCGCTTTCTCTATCAAATCTATAGTCTGCTTGATCAAAAACGAGGATGGAACATATTCTTTCATATTCAAATGTAAATTAGCAACCTCGAATGCTAAATTCATCCCAGGCGAGAAGTTCATTGAGACAAAGGCTGATGGGAGTAAGGTGGAAGCATTGATAACGTTTGAAGGGAACAAGATGACACACATACAGATAGAGAGTAACGGCAGAACTTCGACACATTTACGCGAATTCTTCCCCGATAGACTTATCGTGACAACTACGGCTAAAGGATTTGATAAAATTGTTACCAGGAAATTTGAACTGGTCAAGGTGGTTTCGGGTGGTGGTGTTTGGGGGGAAAGGGACGCTAGCTGTGGGAGGGGGGGAGGAGGGCTGGTGTCGTTGTCATGGAAACGCGGCGCCCGGACGCCAGATCCCGACGCCCGCAGTGTCGGTCCCCAGCCGCCCGCAGCCCCTACCCCCGCCGCACAAGTGACAAGCGCTGCGAAAAAGCACAACCGCCCTGTGTACGGGGACTATGATTTTTGGGAAACACGAATTTTGTAA
>DPOGS207013-PA
MDNFLGKEYVMVSQENFDDYLVFMGVSYLNRKLAFSIKSIVCLIKNEDGTYSFIFKCKLATSNAKFIPGEKFIETKADGSKVEALITFEGNKMTHIQIESNGRTSTHLREFFPDRLIVTTTAKGFDKIVTRKFELVKVVSGGGVWGERDASCGRGGGGLVSLSWKRGARTPDPDARSVGPQPPAAPTPAAQVTSAAKKHNRPVYGDYDFWETRIL-