| DPOGS207689 | ||
|---|---|---|
| Transcript | DPOGS207689-TA | 528 bp |
| Protein | DPOGS207689-PA | 175 aa |
| Genomic position | DPSCF300502 - 29178-30999 | |
| RNAseq coverage | 24x (Rank: top 78%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL003772 | 2e-93 | 86.78% | |
| Bombyx | BGIBMGA000167-TA | 3e-88 | 83.72% | |
| Drosophila | CG1707-PA | 1e-70 | 69.59% | |
| EBI UniRef50 | UniRef50_Q04760 | 4e-64 | 65.71% | Lactoylglutathione lyase n=129 Tax=cellular organisms RepID=LGUL_HUMAN |
| NCBI RefSeq | XP_002017809.1 | 2e-71 | 71.93% | GL17374 [Drosophila persimilis] |
| NCBI nr blastp | gi|195153795 | 3e-70 | 71.93% | GL17374 [Drosophila persimilis] |
| NCBI nr blastx | gi|195123123 | 7e-70 | 74.10% | GI20823 [Drosophila mojavensis] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0004462 | 2.4e-58 | lactoylglutathione lyase activity | |
| GO:0046872 | 2.4e-58 | metal ion binding | ||
| KEGG pathway | dpo:Dpse_GA24807 | 5e-71 | ||
| K01759 (E4.4.1.5, GLO1, gloA) | maps-> | MAPK signaling pathway - yeast | ||
| Pyruvate metabolism | ||||
| InterPro domain | [15-173] IPR004361 | 2.4e-58 | Glyoxalase I | |
| [29-168] IPR004360 | 4.9e-24 | Glyoxalase/fosfomycin resistance/dioxygenase | ||
| Orthology group | MCL12663 | Multiple-copy universal gene | ||
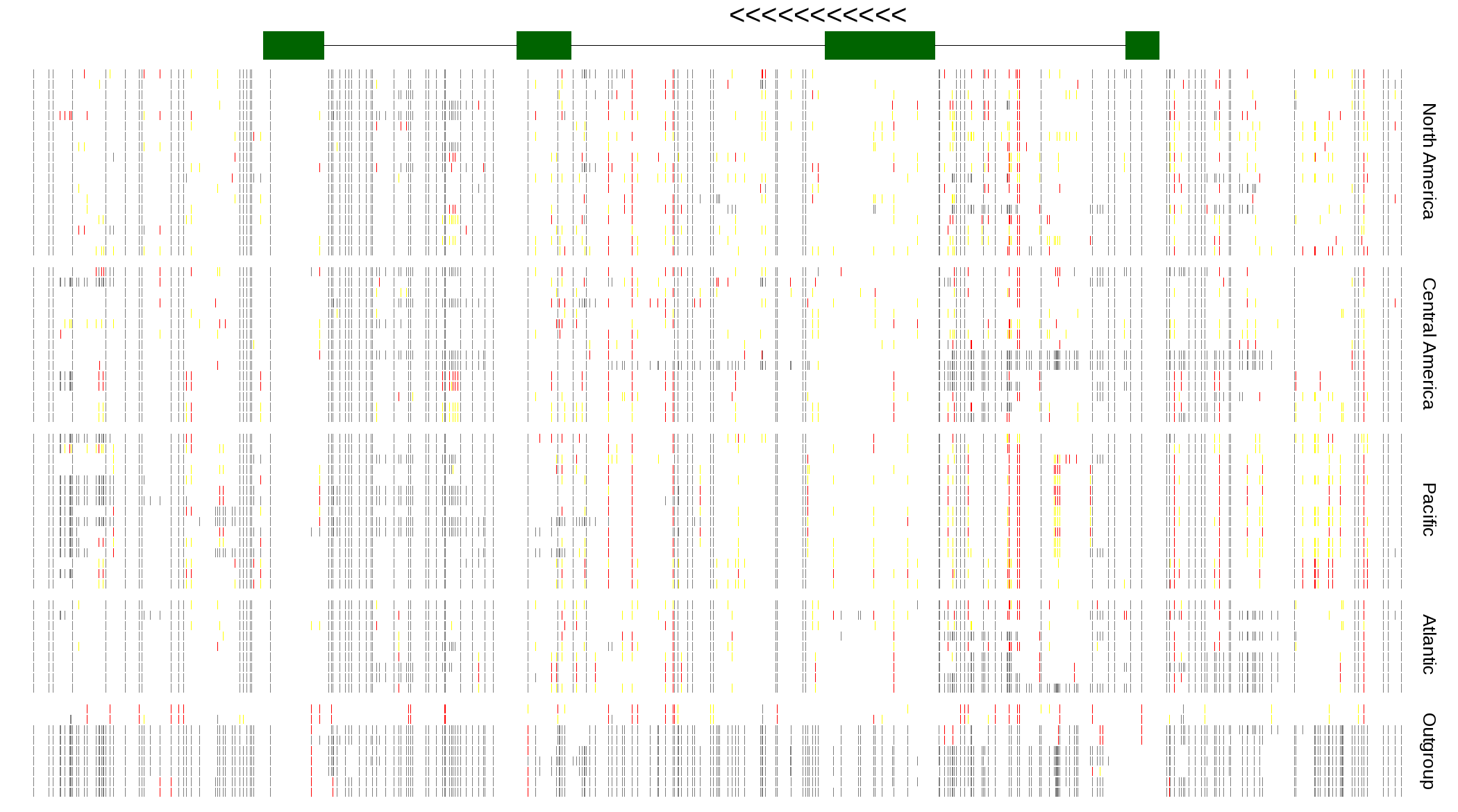
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS207689-TA
ATGGCTAGCGAAGGTTTTTCACCACAGGAGATAGAAGCCCTCTGCCAAACACCTAGTCCTAATACAAAGGATTTTATGTTCCAACAAACCATGTATAGGATAAAGGATCCTCGTAAATCATTACCGTTTTACACCGGAGTCCTCGGGATGACACTGTTGAAGCAATTGCATTTTCCCGAGATGAAATTCTCGTTGTTCTTCATGGGTTACGAAAATCCTAATGAAATACCAAAAGATGAAAAGGAAAGAAGTCAGTGGGCTATGTCTAGAAAGGCCACATTGGAATTGACTTATAATTGGGGCACGGAAAACGATAACACAGTTTATCACAACGGCAACTCAGATCCGAGAGGGTTCGGTCACATCGGTATACTTGTGCCCGATGTTGATGAAGCCTGTGAACGTTTCGAACAACAAGGTGTGAAATTCGTAAAGAGGCCTCAAGATGGAAAAATGAAAGGCCTCGCTTTCATACAGGACCCGGATGGTTACTGGATAGAAATATTCACTAACAAAGTTGTTTCGTAA
>DPOGS207689-PA
MASEGFSPQEIEALCQTPSPNTKDFMFQQTMYRIKDPRKSLPFYTGVLGMTLLKQLHFPEMKFSLFFMGYENPNEIPKDEKERSQWAMSRKATLELTYNWGTENDNTVYHNGNSDPRGFGHIGILVPDVDEACERFEQQGVKFVKRPQDGKMKGLAFIQDPDGYWIEIFTNKVVS-