| DPOGS208035 | ||
|---|---|---|
| Transcript | DPOGS208035-TA | 762 bp |
| Protein | DPOGS208035-PA | 253 aa |
| Genomic position | DPSCF300203 + 42203-45494 | |
| RNAseq coverage | 27712x (Rank: top 0%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL004054 | 3e-133 | 97.23% | |
| Bombyx | BGIBMGA001501-TA | 4e-132 | 95.65% | |
| Drosophila | RpS6-PB | 3e-106 | 78.51% | |
| EBI UniRef50 | UniRef50_P62753 | 6e-100 | 73.71% | 40S ribosomal protein S6 n=462 Tax=root RepID=RS6_HUMAN |
| NCBI RefSeq | NP_001037566.1 | 1e-130 | 95.65% | ribosomal protein S6 [Bombyx mori] |
| NCBI nr blastp | gi|315115403 | 9e-139 | 98.80% | ribosomal protein S6 [Euphydryas aurinia] |
| NCBI nr blastx | gi|342356391 | 1e-135 | 97.23% | ribosomal protein S6 [Heliconius melpomene cythera] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005840 | 5.4e-207 | ribosome | |
| GO:0006412 | 5.4e-207 | translation | ||
| GO:0005622 | 5.4e-207 | intracellular | ||
| GO:0003735 | 5.4e-207 | structural constituent of ribosome | ||
| KEGG pathway | nvi:100118087 | 5e-118 | ||
| K02991 (RP-S6e, RPS6) | maps-> | Ribosome | ||
| Insulin signaling pathway | ||||
| mTOR signaling pathway | ||||
| InterPro domain | [2-249] IPR001377 | 5.4e-207 | Ribosomal protein S6e | |
| [1-251] IPR014401 | 4e-184 | Ribosomal protein S6, eukaryotic | ||
| Orthology group | MCL13190 | Single-copy universal gene | ||
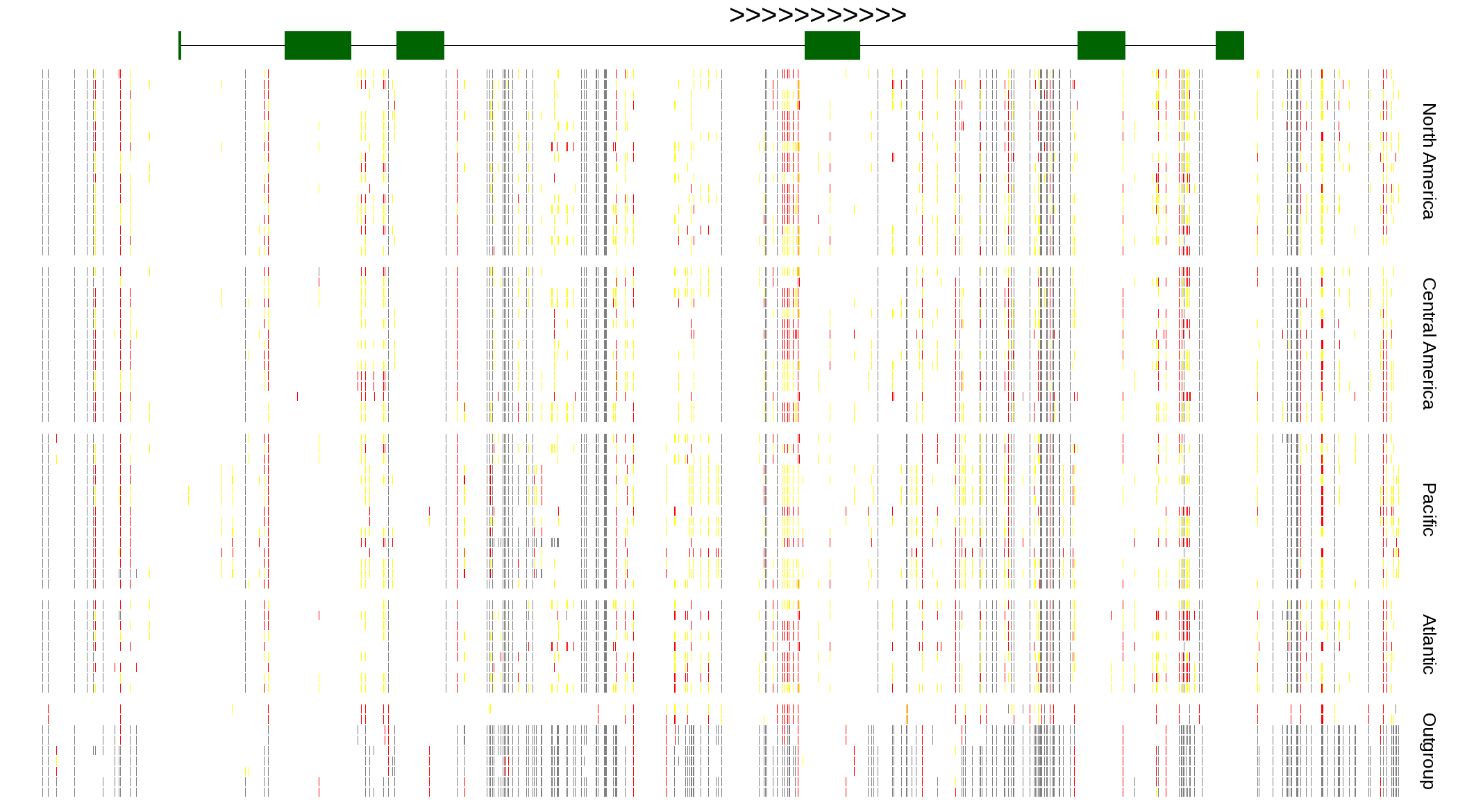
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS208035-TA
ATGAAGTTGAACGTCTCCTACCCGGCAACGGGATGTCAGAAGTTGTTCGAAGTTGTAGATGAGCATAAGCTCCGCATCTTCTATGAGAAGCGCATGGGCGCGGAGGTGGAGGCCGACCAGCTAGGAGATGAATGGAAAGGCTACATCCTCCGCATTGCCGGTGGTAACGACAAACAAGGGTTCCCCATGAAACAGGGTGTCCTTACCAACAGTCGTGTACGTTTGCTGATGTCAAAGGGTCACTCTTGCTACAGACCCCGTCGTGACGGTGAGAGAAAACGCAAGTCGGTTCGTGGATGTATCGTGGACGCCAATCTCTCCGTTTTAGCACTTGTCATTGTTCGTAAAGGAGAACAGGAAATCCCTGGACTCACCGATGGCAATGTCCCCCGTCGTCTCGGACCCAAACGTGCTTCCAAGATCAGGAAGCTCTTCAACCTGACCAAGCAGGATGATGTGCGACGATATGTTGTCAAGCGTCTCCTACCAGCCAAGGAGGGCAAAGAAAACGCTAAACCCAGATATAAGGCCCCTAAAATCCAAAGGTTAGTAACTCCAGTGGTATTGCAACGCAGACGTCACCGATTGGCTCTTAAGAAGAAGCGTCTCGCTAAACGCAAGGCCTCCGAGGCAGAGTATGCTAAACTGCTCGCACAGAGAAAGAAGGAGTCCAAGGTGCGTCGCCAAGAGGAAATCAAACGCAGGCGCTCAGCCTCGATCCGCGACTCGAAGAGTTCCAGCACGAGCGCCCCACAAAAGTGA
>DPOGS208035-PA
MKLNVSYPATGCQKLFEVVDEHKLRIFYEKRMGAEVEADQLGDEWKGYILRIAGGNDKQGFPMKQGVLTNSRVRLLMSKGHSCYRPRRDGERKRKSVRGCIVDANLSVLALVIVRKGEQEIPGLTDGNVPRRLGPKRASKIRKLFNLTKQDDVRRYVVKRLLPAKEGKENAKPRYKAPKIQRLVTPVVLQRRRHRLALKKKRLAKRKASEAEYAKLLAQRKKESKVRRQEEIKRRRSASIRDSKSSSTSAPQK-