| DPOGS209326 | ||
|---|---|---|
| Transcript | DPOGS209326-TA | 342 bp |
| Protein | DPOGS209326-PA | 113 aa |
| Genomic position | DPSCF300234 + 124968-125637 | |
| RNAseq coverage | 1218x (Rank: top 10%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL005870 | 3e-53 | 83.33% | |
| Bombyx | BGIBMGA013879-TA | 6e-33 | 70.45% | |
| Drosophila | CG30016-PA | 6e-20 | 40.74% | |
| EBI UniRef50 | UniRef50_UPI000224652E | 8e-24 | 45.69% | UPI000224652E related cluster n=1 Tax=unknown RepID=UPI000224652E |
| NCBI RefSeq | XP_001608036.1 | 1e-24 | 45.69% | PREDICTED: similar to ENSANGP00000020773 [Nasonia vitripennis] |
| NCBI nr blastp | gi|375143798 | 1e-25 | 50.46% | unnamed protein product [Niastella koreensis GR20-10] |
| NCBI nr blastx | gi|375143798 | 7e-26 | 50.46% | unnamed protein product [Niastella koreensis GR20-10] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0006810 | 2e-39 | transport | |
| KEGG pathway | cqu:CpipJ_CPIJ014057 | 4e-23 | ||
| K07127 (uraH, pucM, hiuH) | maps-> | Purine metabolism | ||
| InterPro domain | [3-113] IPR023416 | 2.4e-40 | Transthyretin/hydroxyisourate hydrolase, superfamily | |
| [5-113] IPR014306 | 2e-39 | Hydroxyisourate hydrolase | ||
| [5-25] IPR000895 | 3.5e-09 | Transthyretin/hydroxyisourate hydrolase | ||
| Orthology group | MCL15246 | Multiple-copy universal gene | ||
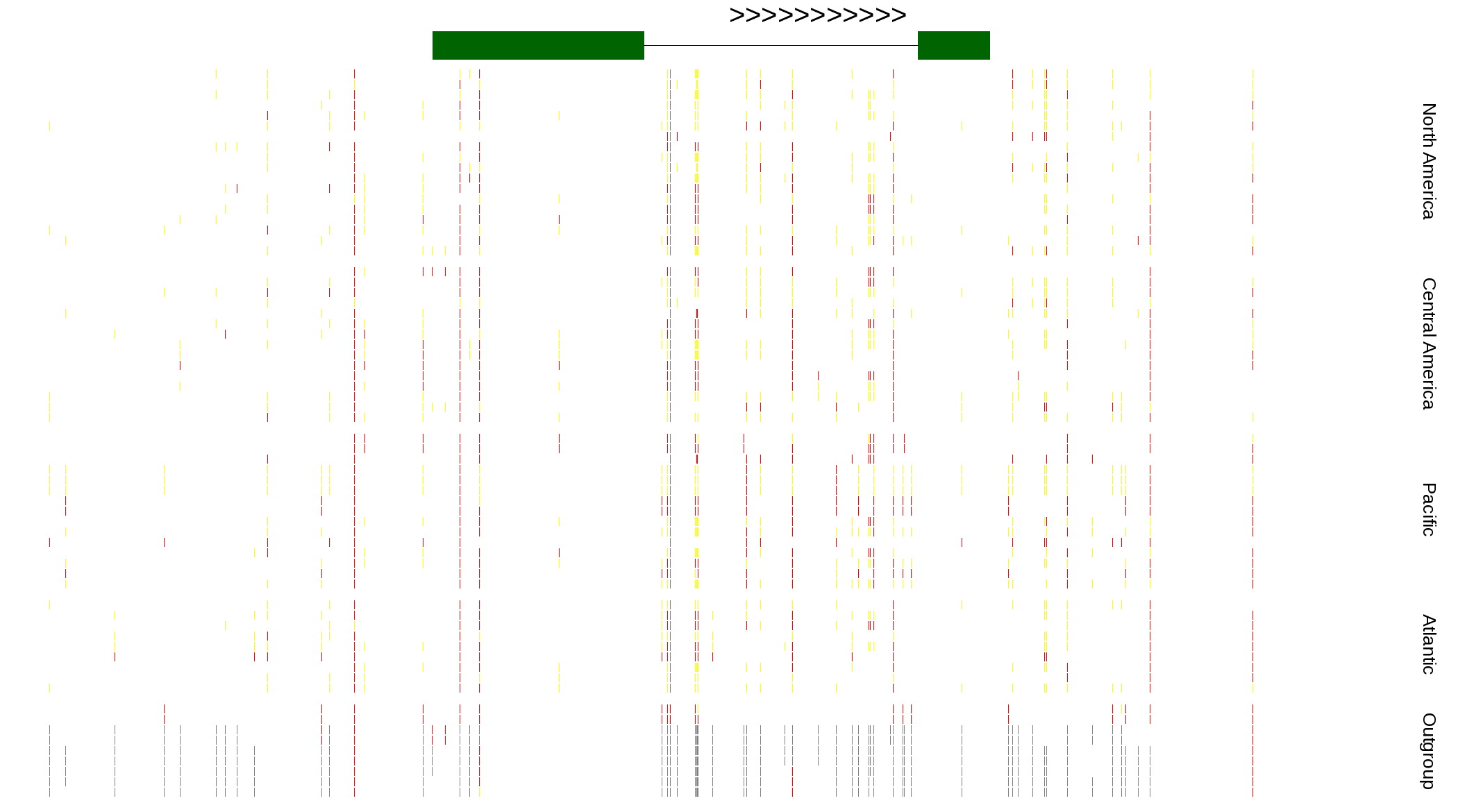
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS209326-TA
ATGTCTCGTCCCGTGTTATCGACGCACGCCCTGGACACTTCCACCGGGAGACCCGTTGTCGGTATTTTTGTGGAGTTGTACAAGAAGAGAGACAACTCGTGGACACAGTGGCACAACACAGTGACATCTAGCGACGGAAGGATACAGTTCCCGTTCTCGAAGGACTCGATGCCCGCCGGCACCTACAAGCTCAAGTTCAACGTCGAGGACTATTACAAGAGACACGACCAGGAGACTCTGTACCCGTTCGTGGAGATCGTGTTCAACACTAAGGAAGACGAACACTACCACATCCCGTTGCTGCTGAGCCCCTTCGGATACTCCACCTACAGAGGCAGCTAG
>DPOGS209326-PA
MSRPVLSTHALDTSTGRPVVGIFVELYKKRDNSWTQWHNTVTSSDGRIQFPFSKDSMPAGTYKLKFNVEDYYKRHDQETLYPFVEIVFNTKEDEHYHIPLLLSPFGYSTYRGS-