| DPOGS209578 | ||
|---|---|---|
| Transcript | DPOGS209578-TA | 714 bp |
| Protein | DPOGS209578-PA | 237 aa |
| Genomic position | DPSCF300015 - 1029427-1032087 | |
| RNAseq coverage | 222x (Rank: top 45%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL017040 | 7e-47 | 43.36% | |
| Bombyx | BGIBMGA002279-TA | 1e-40 | 37.04% | |
| Drosophila | CG16936-PC | 4e-37 | 36.53% | |
| EBI UniRef50 | UniRef50_D2I930 | 6e-64 | 50.91% | Glutathione S-transferase epsilon 2 n=1 Tax=Spodoptera litura RepID=D2I930_SPOLT |
| NCBI RefSeq | XP_002004610.1 | 3e-40 | 39.27% | GI19515 [Drosophila mojavensis] |
| NCBI nr blastp | gi|270313655 | 2e-63 | 50.91% | glutathione S-transferase epsilon 2 [Spodoptera litura] |
| NCBI nr blastx | gi|270313655 | 6e-61 | 50.91% | glutathione S-transferase epsilon 2 [Spodoptera litura] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005515 | 2.6e-07 | protein binding | |
| KEGG pathway | dpo:Dpse_GA14540 | 3e-34 | ||
| K00799 (E2.5.1.18, gst) | maps-> | Drug metabolism - cytochrome P450 | ||
| Glutathione metabolism | ||||
| Metabolism of xenobiotics by cytochrome P450 | ||||
| InterPro domain | [97-223] IPR010987 | 1.1e-23 | Glutathione S-transferase, C-terminal-like | |
| [13-101] IPR012336 | 5.5e-22 | Thioredoxin-like fold | ||
| [28-87] IPR004045 | 2.6e-07 | Glutathione S-transferase, N-terminal | ||
| Orthology group | MCL34807 | Lepidoptera specific | ||
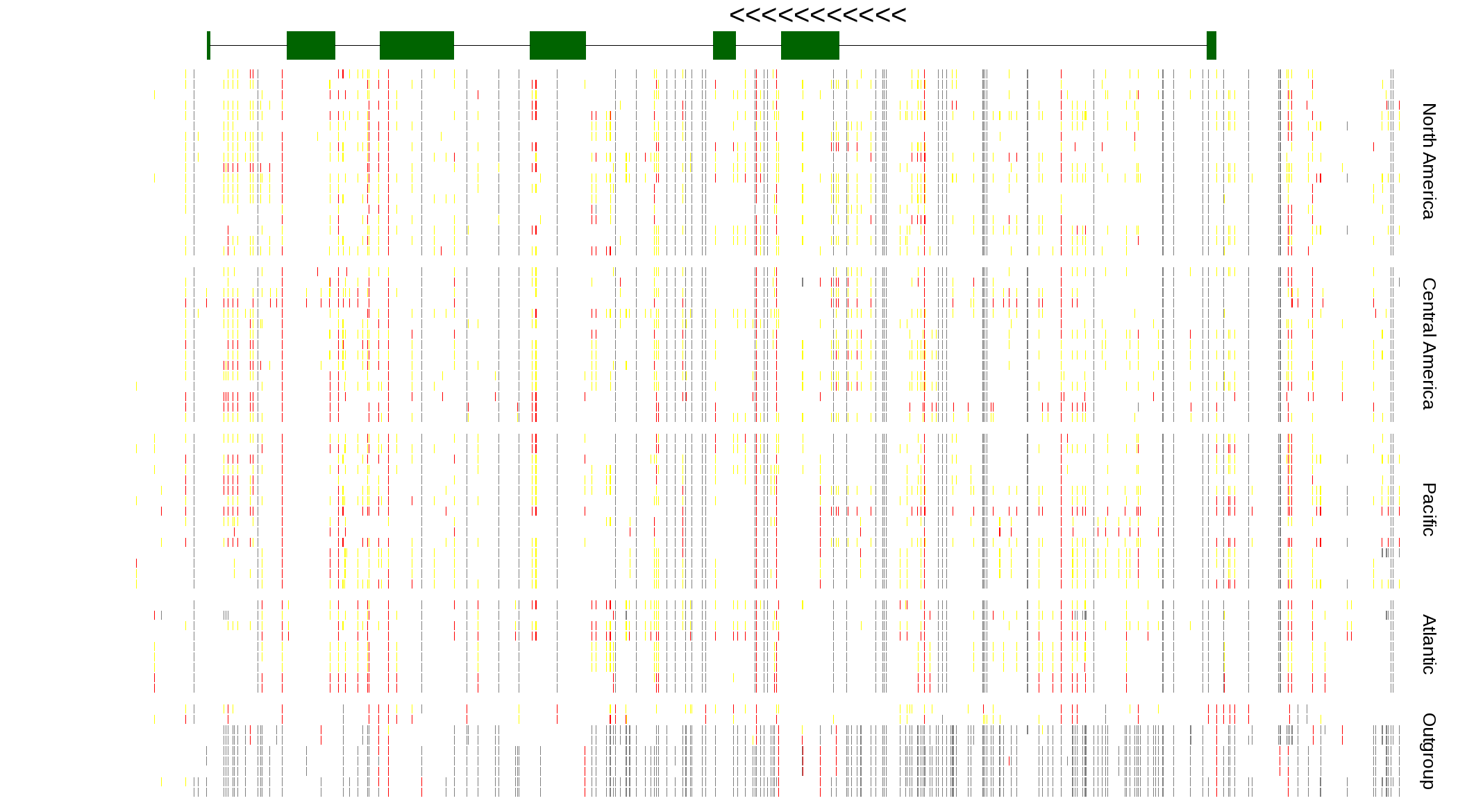
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS209578-TA
ATGTCAAAAGGTAGGTTGAATGAAGTAACAAAGTTACCCCGACTGCTCCTGTACAAGAGAAATGCCAGTCCACCGTCCAGCGCTGTCATGATTCTAGGTGATATGCTAGGATTAAATTTCGACTATAGGGAACCTGATTTAATAAAACTGGAGCATCGATCTCCAGAATTTAAGAAGATAAATCCCATGGCAACCATCCTAGTGCTTCAGGATGGAGATGTAACTATATGCGAAAGTCACGCAATTATGAAATACCTAGTAAATAAATATGGTGGTGAGAGATGTGAAAGGCTTTATCCCGCTGATTTGAGTGTCCGAGCTAACATTGATCAACTAATGTTCTATGATGCTGGGGTCTTGTTTGTTAGGCTTAAGGTTGTCGCGCTACCAACAATGCTTCAAGGCTTAACAGGTCCAACCAAAGAGCAAGTTGCAGATATTGATGAAGGTTACACAGTACTCGAGGCCTATTTAAATAAACACTCGTATATAGCAACTGATCATTTAACTATAGCTGATTTATCTGTTGGCACAACAACTACAGCATTGCAGTCAGTACACAAGCTAGATAAAAATAGGTTTCCTCTAAGTGCAGAATGGCTGGAACGATTAAAGGGTGAAAAGTCTTTCAAAAAATTTAATGAACCTAGTGTAAAAGAATTATCCACAATATTAAACGTATTTTGGAAGAAAAATAAGGAGCGAATCAGATGA
>DPOGS209578-PA
MSKGRLNEVTKLPRLLLYKRNASPPSSAVMILGDMLGLNFDYREPDLIKLEHRSPEFKKINPMATILVLQDGDVTICESHAIMKYLVNKYGGERCERLYPADLSVRANIDQLMFYDAGVLFVRLKVVALPTMLQGLTGPTKEQVADIDEGYTVLEAYLNKHSYIATDHLTIADLSVGTTTTALQSVHKLDKNRFPLSAEWLERLKGEKSFKKFNEPSVKELSTILNVFWKKNKERIR-