| DPOGS209737 | ||
|---|---|---|
| Transcript | DPOGS209737-TA | 459 bp |
| Protein | DPOGS209737-PA | 152 aa |
| Genomic position | DPSCF300105 + 340388-341245 | |
| RNAseq coverage | 185x (Rank: top 49%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL011350 | 1e-68 | 74.83% | |
| Bombyx | BGIBMGA008925-TA | 1e-57 | 75.56% | |
| Drosophila | CG8353-PA | 5e-32 | 41.03% | |
| EBI UniRef50 | UniRef50_Q2F6C1 | 1e-64 | 75.50% | Cytidine deaminase n=2 Tax=Obtectomera RepID=Q2F6C1_BOMMO |
| NCBI RefSeq | NP_001040110.1 | 3e-65 | 75.50% | cytidine deaminase [Bombyx mori] |
| NCBI nr blastp | gi|114051311 | 5e-64 | 75.50% | cytidine deaminase [Bombyx mori] |
| NCBI nr blastx | gi|114051311 | 5e-61 | 75.50% | cytidine deaminase [Bombyx mori] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0009972 | 1.9e-35 | cytidine deamination | |
| GO:0004126 | 1.9e-35 | cytidine deaminase activity | ||
| GO:0003824 | 2.6e-35 | catalytic activity | ||
| GO:0016787 | 1.2e-16 | hydrolase activity | ||
| GO:0008270 | 1.2e-16 | zinc ion binding | ||
| KEGG pathway | aag:AaeL_AAEL006518 | 1e-33 | ||
| K01489 (E3.5.4.5, cdd) | maps-> | Drug metabolism - other enzymes | ||
| Pyrimidine metabolism | ||||
| InterPro domain | [18-144] IPR006262 | 1.9e-35 | Cytidine deaminase, homotetrameric | |
| [12-152] IPR016193 | 2.6e-35 | Cytidine deaminase-like | ||
| [15-112] IPR002125 | 1.2e-16 | CMP/dCMP deaminase, zinc-binding | ||
| Orthology group | MCL12324 | Single-copy universal gene | ||
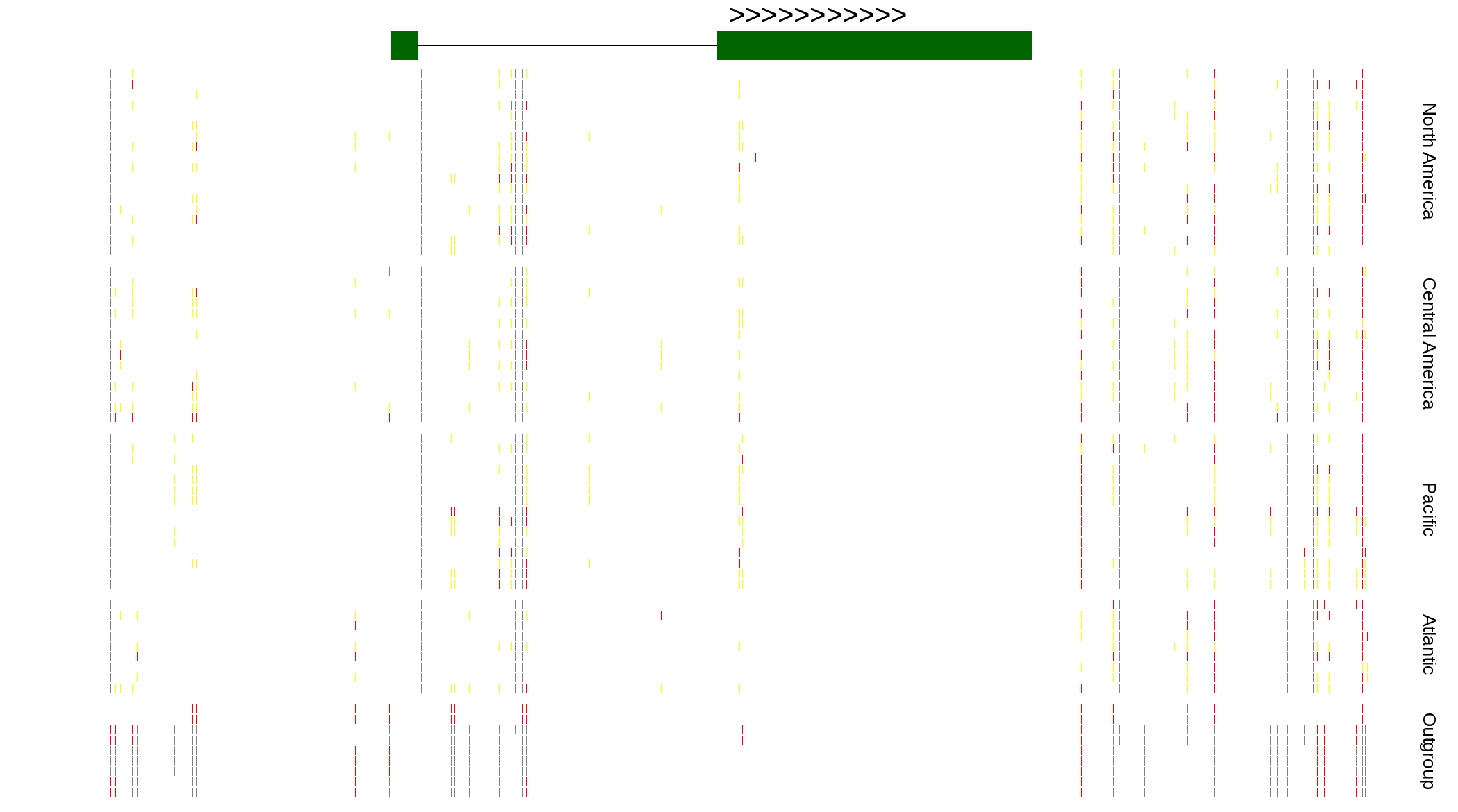
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS209737-TA
ATGGAGCAATATGATATAGTGGACTTCTCCTCATTAGGTGAAACAGCAAAACAATTAATACAAGAGGCGGCACACATACGACAACGGGCTTACTGTCCGTATTCCAACTTCGCAGTGGGAGCCGCAATCCTTACAGAAGAGGACGAACGAATGTACACGGGATGCAACATAGAAAGCTCGACACTCACTCCTACTATATGCGCGGAAAGAGCCGCGGTTCCGAGGGCTGTTTGTGACGGCTACACCAAATTCAAAATGGTAGCAGTGGTGGCACATCAGAAGGAAACATTCACCGCCCCCTGTGGCGTCTGCAGGCAGGTCCTGAGCGAGTTCCGCAGCTCAGATGGAGACATCGAAATCTATCTCAGCAAACCCACTATGGACAAAATCTTGTGCACCAAAATGTCTAAACTTTTACCACTTTCATTCGTGAGTTACAAAATAGACAGTGTGACATAA
>DPOGS209737-PA
MEQYDIVDFSSLGETAKQLIQEAAHIRQRAYCPYSNFAVGAAILTEEDERMYTGCNIESSTLTPTICAERAAVPRAVCDGYTKFKMVAVVAHQKETFTAPCGVCRQVLSEFRSSDGDIEIYLSKPTMDKILCTKMSKLLPLSFVSYKIDSVT-