| DPOGS210004 | ||
|---|---|---|
| Transcript | DPOGS210004-TA | 432 bp |
| Protein | DPOGS210004-PA | 143 aa |
| Genomic position | DPSCF300327 - 82759-84029 | |
| RNAseq coverage | 948x (Rank: top 13%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL009871 | 2e-66 | 82.64% | |
| Bombyx | BGIBMGA008414-TA | 5e-43 | 80.41% | |
| Drosophila | CG3719-PA | 2e-33 | 51.47% | |
| EBI UniRef50 | UniRef50_Q16V30 | 1e-36 | 56.30% | Thioredoxin n=3 Tax=Culicidae RepID=Q16V30_AEDAE |
| NCBI RefSeq | XP_974436.2 | 3e-40 | 67.80% | PREDICTED: similar to thioredoxin (AGAP003338-PA) [Tribolium castaneum] |
| NCBI nr blastp | gi|189236262 | 8e-39 | 67.80% | PREDICTED: similar to thioredoxin (AGAP003338-PA) [Tribolium castaneum] |
| NCBI nr blastx | gi|332375018 | 5e-40 | 65.38% | unknown [Dendroctonus ponderosae] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0015035 | 2.8e-30 | protein disulfide oxidoreductase activity | |
| GO:0009055 | 2.8e-30 | electron carrier activity | ||
| GO:0006662 | 2.8e-30 | glycerol ether metabolic process | ||
| GO:0045454 | 2.8e-30 | cell redox homeostasis | ||
| KEGG pathway | ||||
| InterPro domain | [24-135] IPR005746 | 2.8e-30 | Thioredoxin | |
| [30-138] IPR012336 | 4.8e-28 | Thioredoxin-like fold | ||
| [36-133] IPR013766 | 5.4e-20 | Thioredoxin domain | ||
| Orthology group | MCL16500 | Insect specific | ||
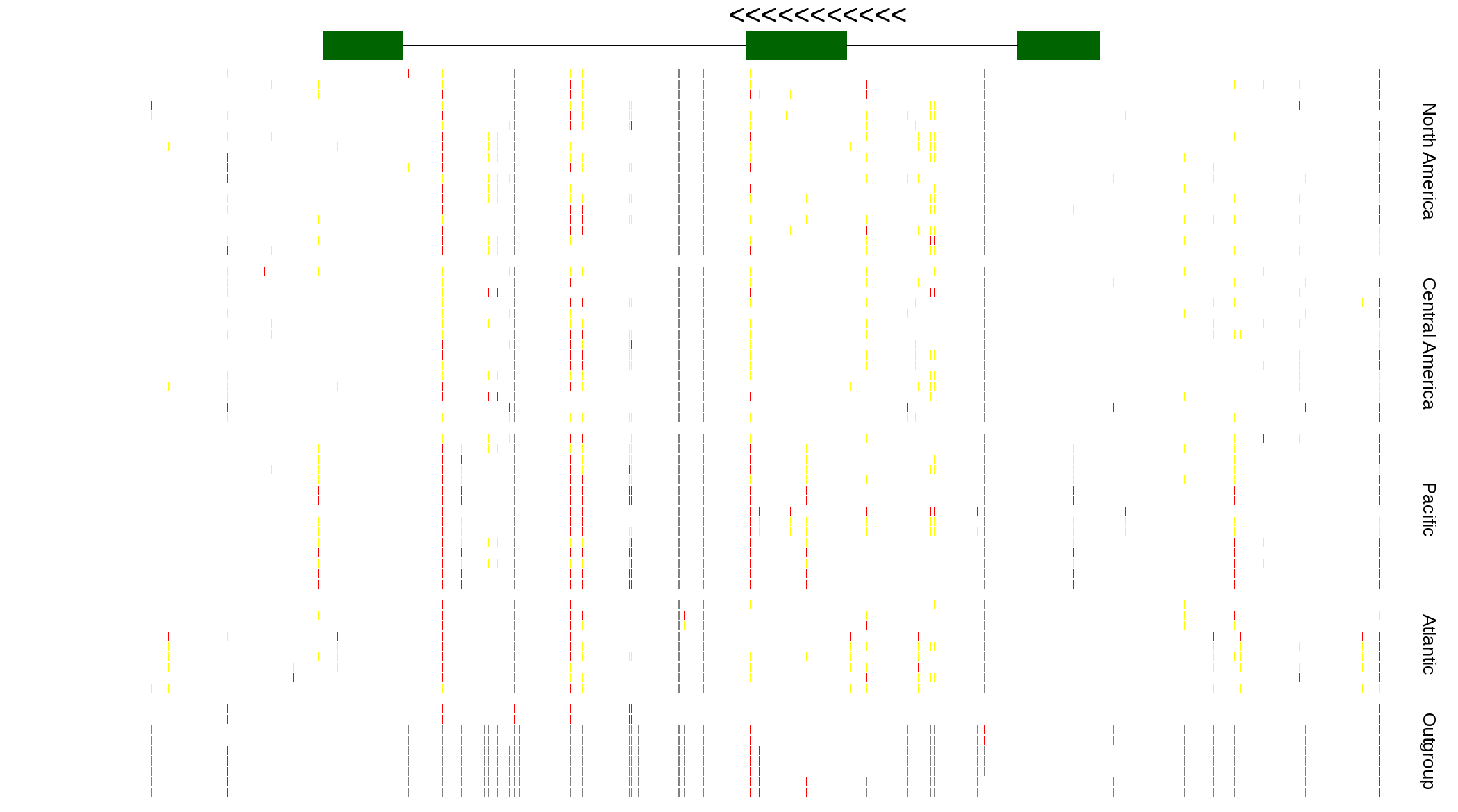
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS210004-TA
ATGAATAATGCCATCTGTCAAGGACTGCGGTCACCAGTTCGGAGGCTCGCTGGTCTAAGAAATAAACTTATATCAACATCTGTTGTCCACAATGAGACCATTCTAGTCAAGAATTACGATGATTTCGTAAATAAGGTGATGAATAACGAGAAACCTGTGATAGTGAACTTCCACGCGGAGTGGTGTGAGCCGTGCAAGATCCTCACGCCGAAGCTCAAGGAGTTGGTTGAACCGTTCACTTCGCTGGATCTCGCCATAGTCGACGTGGAAGACAACGCGGAGCTAGTTCACACTTTTGAGGTGAAAGCGGTACCGGCGGTTATAGCCATCAGGAATAGTCTCATAGTGGACAAATTCATCGGCCTTGTGGACACGGACATGATCAACAGTCTCATAGAACGGATGGCGGGGAAACGGAATAATGGCGCGTAA
>DPOGS210004-PA
MNNAICQGLRSPVRRLAGLRNKLISTSVVHNETILVKNYDDFVNKVMNNEKPVIVNFHAEWCEPCKILTPKLKELVEPFTSLDLAIVDVEDNAELVHTFEVKAVPAVIAIRNSLIVDKFIGLVDTDMINSLIERMAGKRNNGA-