| DPOGS210102 | ||
|---|---|---|
| Transcript | DPOGS210102-TA | 735 bp |
| Protein | DPOGS210102-PA | 244 aa |
| Genomic position | DPSCF300017 + 771309-776867 | |
| RNAseq coverage | 10563x (Rank: top 1%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL002978 | 4e-45 | 66.67% | |
| Bombyx | BGIBMGA002788-TA | 7e-28 | 37.82% | |
| Drosophila | pk-PC | 6e-26 | 33.12% | |
| EBI UniRef50 | UniRef50_E3WVE2 | 2e-102 | 63.09% | Putative uncharacterized protein n=1 Tax=Anopheles darlingi RepID=E3WVE2_ANODA |
| NCBI RefSeq | XP_001688656.1 | 1e-110 | 77.05% | AGAP005400-PB [Anopheles gambiae str. PEST] |
| NCBI nr blastp | gi|158294126 | 2e-109 | 77.05% | AGAP005400-PB [Anopheles gambiae str. PEST] |
| NCBI nr blastx | gi|158294126 | 1e-115 | 77.05% | AGAP005400-PB [Anopheles gambiae str. PEST] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008270 | 6.5e-18 | zinc ion binding | |
| KEGG pathway | tca:658324 | 7e-26 | ||
| K05760 (PXN) | maps-> | Chemokine signaling pathway | ||
| Regulation of actin cytoskeleton | ||||
| Leukocyte transendothelial migration | ||||
| Bacterial invasion of epithelial cells | ||||
| Focal adhesion | ||||
| VEGF signaling pathway | ||||
| InterPro domain | [130-186] IPR001781 | 6.5e-18 | Zinc finger, LIM-type | |
| Orthology group | MCL11109 | Multiple-copy universal gene | ||
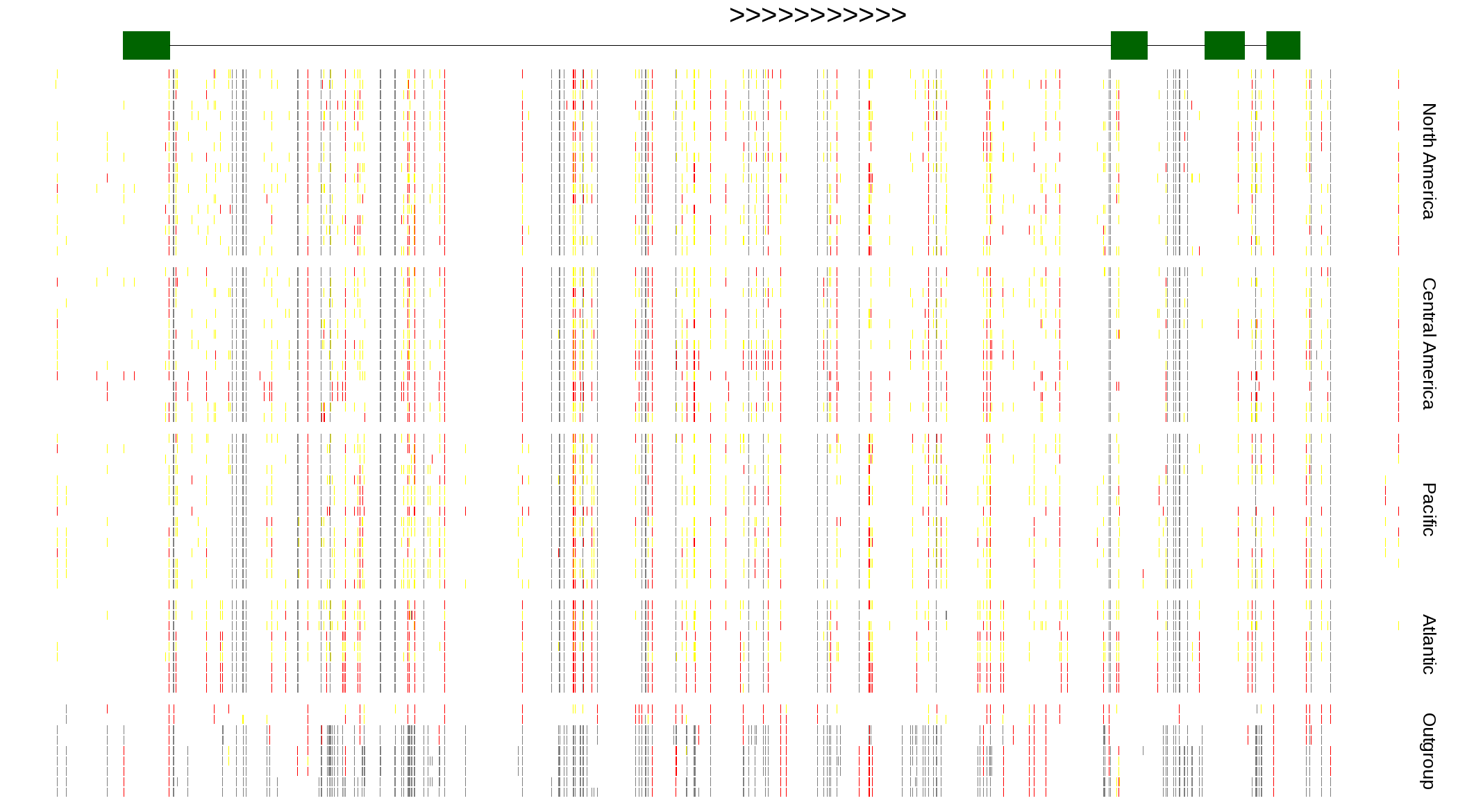
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS210102-TA
ATGTCAGCGGATGTGTTGAGTGATGATTTCAAATCCCGTCTTCATTTGACGACGAAGACTGTTGGACAGGAGAGGATAAGGCGAGCGCTTGAGAACAACGAGGATGTGGCGATACTGTCCATGTTCCTACCGAATGCAACAGCTGAGAGGGAGAATAAAAAATTCTGTTGTGTCATGAAAGACCAATGCCTGCTCGGGGATCCCAAAAAAACTATCGCAGGCACCAAGAAGATGGAGTACAAGACTCGACAGTGGCATGAGAAGTGTTTCTGCTGCGTGGTGTGCAAGAAAGCCATTGGGACCAAGAGCTTCATCCCTCGGGAACAGGAGATCTACTGCACTGGTTGTTACGAGAATAAGTTCTCTACGCGCTGCGTCAAATGCAATAAGATTATAACGCAAGACGGCGTGACGTACAAGCACGAGCCCTGGCACCGCGAGTGCTTCACCTGCACACACTGCAACAACTCGCTGGCCGGGGAGCGCTTCACCTCCAGAGACGAGAAGCCATACTGCAGCGAATGTTTCGGGGAACTGTTCGCCAAGAGATGCACCGCATGCAGCAAGCCCATCACCGGTATCGGTGGCACGCGTTTCATCTCGTTCGAGGATCGTCATTGGCACAACAACTGCTTCCAGTGCGCTTACTGCAAGGTGTCTCTCGTCGGGAAGGGATTCATCACTGTGGAACAGGACGTCATTTGCCCCGAGTGCGCCAAGCAGAAGATCGTGTAA
>DPOGS210102-PA
MSADVLSDDFKSRLHLTTKTVGQERIRRALENNEDVAILSMFLPNATAERENKKFCCVMKDQCLLGDPKKTIAGTKKMEYKTRQWHEKCFCCVVCKKAIGTKSFIPREQEIYCTGCYENKFSTRCVKCNKIITQDGVTYKHEPWHRECFTCTHCNNSLAGERFTSRDEKPYCSECFGELFAKRCTACSKPITGIGGTRFISFEDRHWHNNCFQCAYCKVSLVGKGFITVEQDVICPECAKQKIV-