| DPOGS210477 | ||
|---|---|---|
| Transcript | DPOGS210477-TA | 540 bp |
| Protein | DPOGS210477-PA | 179 aa |
| Genomic position | DPSCF300062 + 500580-501357 | |
| RNAseq coverage | 265x (Rank: top 40%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL021589 | 2e-84 | 76.54% | |
| Bombyx | BGIBMGA002222-TA | 4e-39 | 46.93% | |
| Drosophila | GstD3-PA | 6e-45 | 50.60% | |
| EBI UniRef50 | UniRef50_A8CA00 | 6e-55 | 55.06% | Glutathione S-transferase-like protein n=8 Tax=Endopterygota RepID=A8CA00_PHLPP |
| NCBI RefSeq | NP_001040130.1 | 3e-72 | 68.72% | glutathione S-transferase unclassified 2 [Bombyx mori] |
| NCBI nr blastp | gi|218176145 | 8e-74 | 72.07% | glutathione S-transferase 5 [Choristoneura fumiferana] |
| NCBI nr blastx | gi|218176145 | 1e-73 | 72.47% | glutathione S-transferase 5 [Choristoneura fumiferana] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005515 | 3.1e-10 | protein binding | |
| KEGG pathway | dme:Dmel_CG4381 | 3e-43 | ||
| K00799 (E2.5.1.18, gst) | maps-> | Drug metabolism - cytochrome P450 | ||
| Glutathione metabolism | ||||
| Metabolism of xenobiotics by cytochrome P450 | ||||
| InterPro domain | [49-177] IPR010987 | 1e-32 | Glutathione S-transferase, C-terminal-like | |
| [1-52] IPR012336 | 3.5e-16 | Thioredoxin-like fold | ||
| [84-155] IPR004046 | 2.4e-10 | Glutathione S-transferase, C-terminal | ||
| [3-41] IPR004045 | 3.1e-10 | Glutathione S-transferase, N-terminal | ||
| Orthology group | MCL19715 | Insect specific | ||
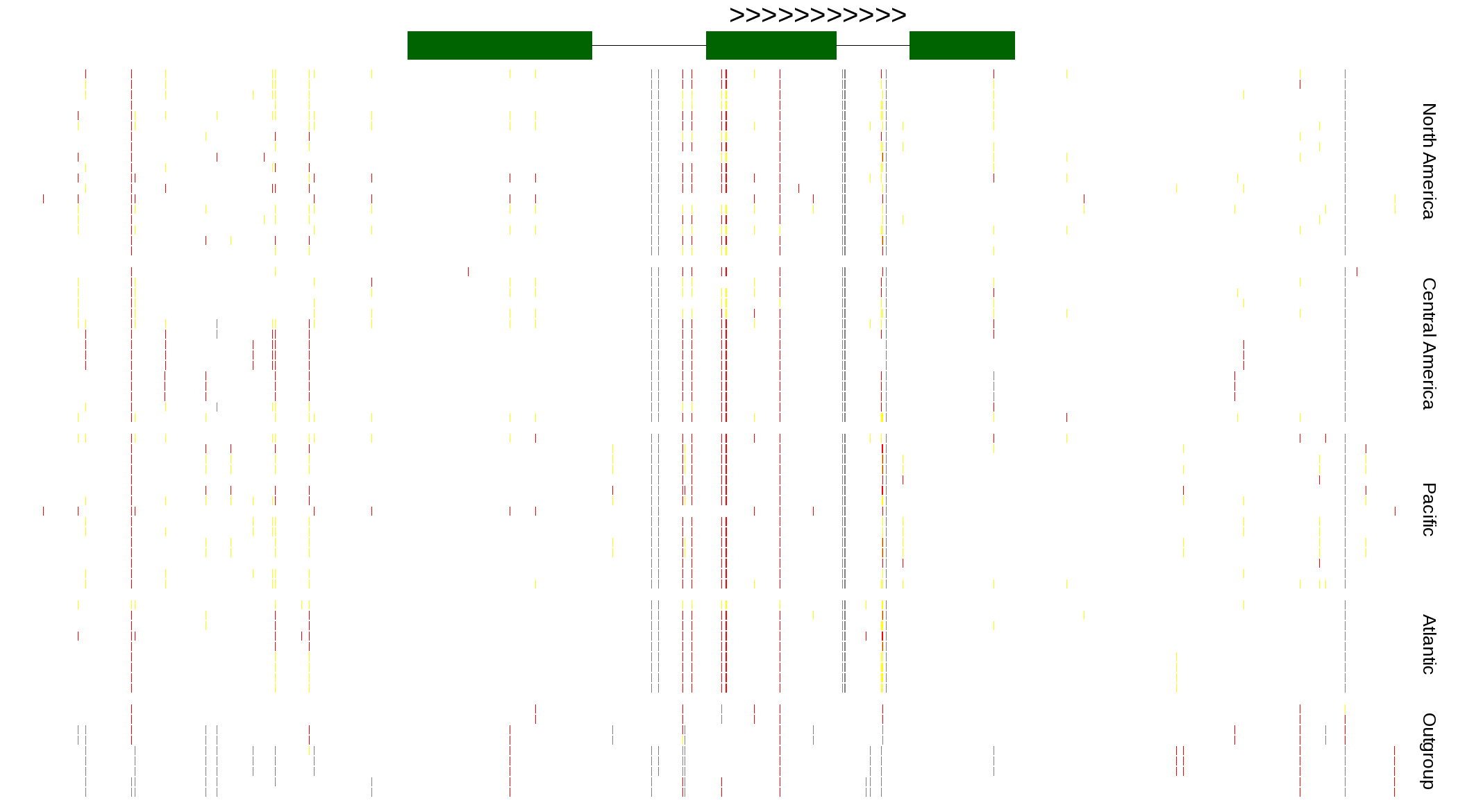
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS210477-TA
ATGAAAAAAGAACAACTGAAAGAATCTTTCATCAAAATAAATCCTCAACATTGTGTGCCAACACTTGAAGATAACGGTTTTGTGATATGGGAAAGCCGGGCAATAGCCTGCTATTTGGCTGATAAATACGGTAAAAACGACGATCACTACCCGAAAGATCTTCAACGCCGTGCTATTGTAAACTCAAGACTTTATTTCGATAGTTCATTTTTGTACCCCAGACTCAGGGCTATTTGTTACCCTATTTTATTTGAAGGCGTGAAAGAAATTAAGGATAAATTAAGAAATGATTTGAATGCGACACTTGGTTTCTTAAATCAATTTTTGGAAGACCAGAAGTGGGTTGCTGGTGACAAGCCCACTATAGCAGATACAGCCATTCTAGCATCCTTTAGTTCTGTAGTTGAACTTGGTTGGGATCTTAGCCAATTCCCTAATATTGAGAGATGGTACAAAGAATGCAGTATTCTGCCAGGTTATGAAGAGAATCTTGAGGGAGCTAGAGCATTTGCCGGATTGGTTAAGAGGAACTTAAAATAA
>DPOGS210477-PA
MKKEQLKESFIKINPQHCVPTLEDNGFVIWESRAIACYLADKYGKNDDHYPKDLQRRAIVNSRLYFDSSFLYPRLRAICYPILFEGVKEIKDKLRNDLNATLGFLNQFLEDQKWVAGDKPTIADTAILASFSSVVELGWDLSQFPNIERWYKECSILPGYEENLEGARAFAGLVKRNLK-