| DPOGS211568 | ||
|---|---|---|
| Transcript | DPOGS211568-TA | 447 bp |
| Protein | DPOGS211568-PA | 148 aa |
| Genomic position | DPSCF300084 - 464832-465955 | |
| RNAseq coverage | 109x (Rank: top 60%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL002527 | 6e-47 | 90.72% | |
| Bombyx | BGIBMGA006507-TA | 3e-46 | 96.67% | |
| Drosophila | Gdh-PA | 4e-45 | 94.44% | |
| EBI UniRef50 | UniRef50_P54385 | 4e-43 | 94.44% | Glutamate dehydrogenase, mitochondrial n=129 Tax=cellular organisms RepID=DHE3_DROME |
| NCBI RefSeq | NP_001040245.1 | 7e-45 | 96.67% | glutamate dehydrogenase [Bombyx mori] |
| NCBI nr blastp | gi|114052462 | 1e-43 | 96.67% | glutamate dehydrogenase [Bombyx mori] |
| NCBI nr blastx | gi|114052462 | 6e-41 | 96.67% | glutamate dehydrogenase [Bombyx mori] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005488 | 1.7e-20 | binding | |
| GO:0006810 | 2.8e-11 | transport | ||
| GO:0016021 | 2.8e-11 | integral to membrane | ||
| KEGG pathway | nvi:100123602 | 1e-43 | ||
| K00261 (E1.4.1.3) | maps-> | Nitrogen metabolism | ||
| Arginine and proline metabolism | ||||
| Alanine, aspartate and glutamate metabolism | ||||
| D-Glutamine and D-glutamate metabolism | ||||
| Proximal tubule bicarbonate reclamation | ||||
| InterPro domain | [45-146] IPR016040 | 1.7e-20 | NAD(P)-binding domain | |
| [2-62] IPR009038 | 2.8e-11 | GOLD | ||
| Orthology group | ||||
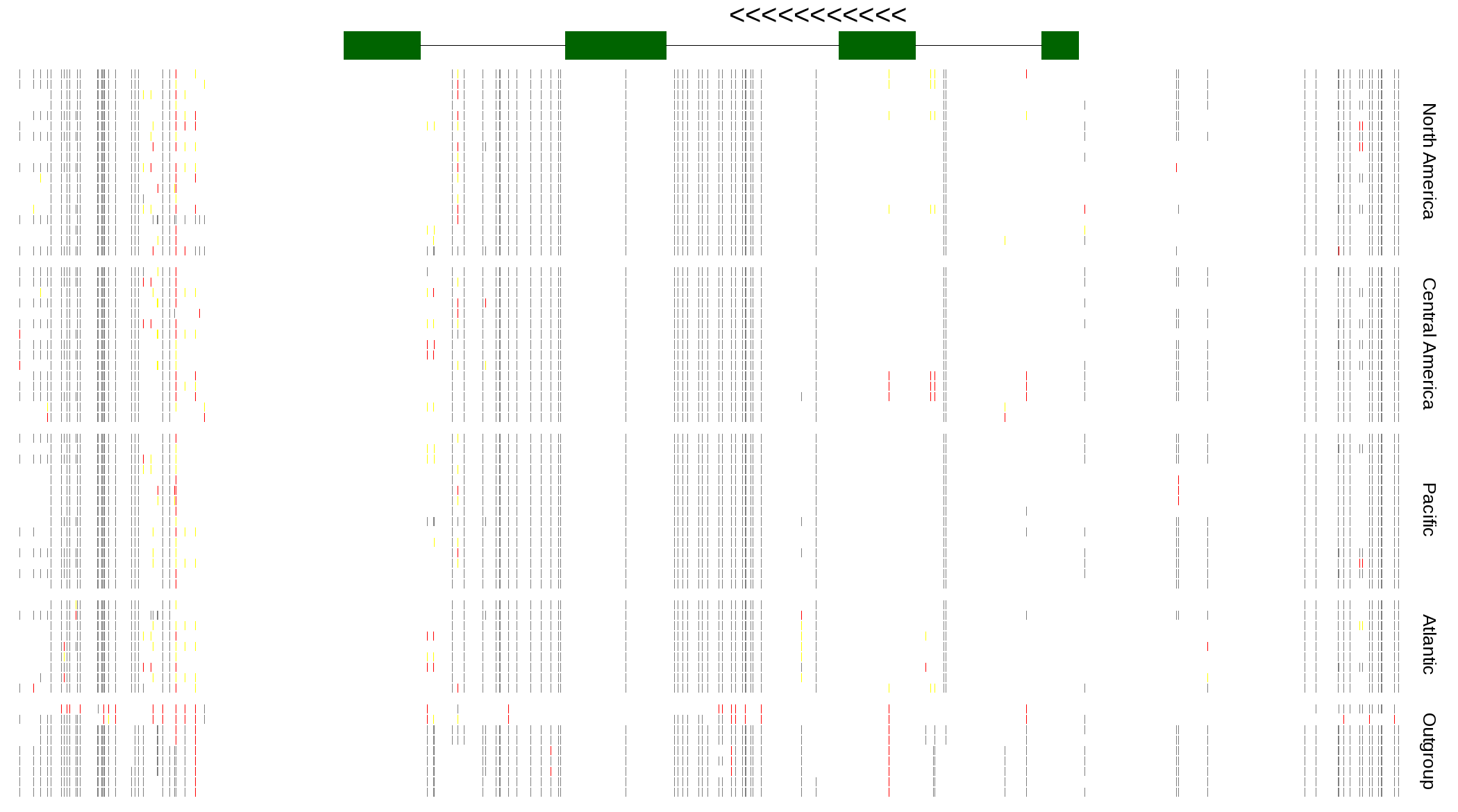
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS211568-TA
ATGGAATGTTTTTATCAGGATATTGAACAAAATACCTCAGCCTCGCTGGAATATCAGGTTATTACTGGAGGCCAGTATGACGTTGATGTGAAAATTGAGGGTCCTAATAAACAAATAATATATCAGCAACAGAAAATGCAATATGACTCTCATCAATTTACAGCACAACACACAGAGTCAGTCCAAGAGTCGTTGGAGCGTCGTTTCGGTCGTGTCGGAGGTCGTATCCCCGTCACAGCCTCCGAGGCCTTCCAGAAGAGGATCTCAGGAGCCTCCGAGAAAGACATCGTCCACTCAGGATTGGACTACACCATGGAAAGATCCGCAAGGGCCATAATGAAAACTGCCATGAAATTCAACCTCGGTCTCGACCTCAGGACGGCGGCGTACGCCAACTCCATCGAAAAAATCTTCACAACCTACGCCGACGCCGGCCTCGCCTTCTAA
>DPOGS211568-PA
MECFYQDIEQNTSASLEYQVITGGQYDVDVKIEGPNKQIIYQQQKMQYDSHQFTAQHTESVQESLERRFGRVGGRIPVTASEAFQKRISGASEKDIVHSGLDYTMERSARAIMKTAMKFNLGLDLRTAAYANSIEKIFTTYADAGLAF-