| DPOGS212228 | ||
|---|---|---|
| Transcript | DPOGS212228-TA | 486 bp |
| Protein | DPOGS212228-PA | 161 aa |
| Genomic position | DPSCF300263 + 10538-11399 | |
| RNAseq coverage | 2189x (Rank: top 6%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL007962 | 2e-82 | 87.97% | |
| Bombyx | BGIBMGA004441-TA | 3e-79 | 83.85% | |
| Drosophila | % | |||
| EBI UniRef50 | UniRef50_E9G2S1 | 1e-24 | 40.46% | Putative uncharacterized protein n=1 Tax=Daphnia pulex RepID=E9G2S1_DAPPU |
| NCBI RefSeq | XP_002737435.1 | 5e-25 | 39.23% | PREDICTED: dual specificity phosphatase 23-like [Saccoglossus kowalevskii] |
| NCBI nr blastp | gi|321475499 | 5e-24 | 40.46% | hypothetical protein DAPPUDRAFT_208196 [Daphnia pulex] |
| NCBI nr blastx | gi|291235004 | 1e-22 | 39.23% | PREDICTED: dual specificity phosphatase 23-like [Saccoglossus kowalevskii] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008138 | 1.4e-10 | protein tyrosine/serine/threonine phosphatase activity | |
| GO:0006470 | 1.4e-10 | protein dephosphorylation | ||
| KEGG pathway | lbc:LACBIDRAFT_239321 | 6e-11 | ||
| K06639 (CDC14) | maps-> | Meiosis - yeast | ||
| Cell cycle - yeast | ||||
| Cell cycle | ||||
| InterPro domain | [10-117] IPR000340 | 1.4e-10 | Dual specificity phosphatase, catalytic domain | |
| Orthology group | MCL18828 | Patchy | ||
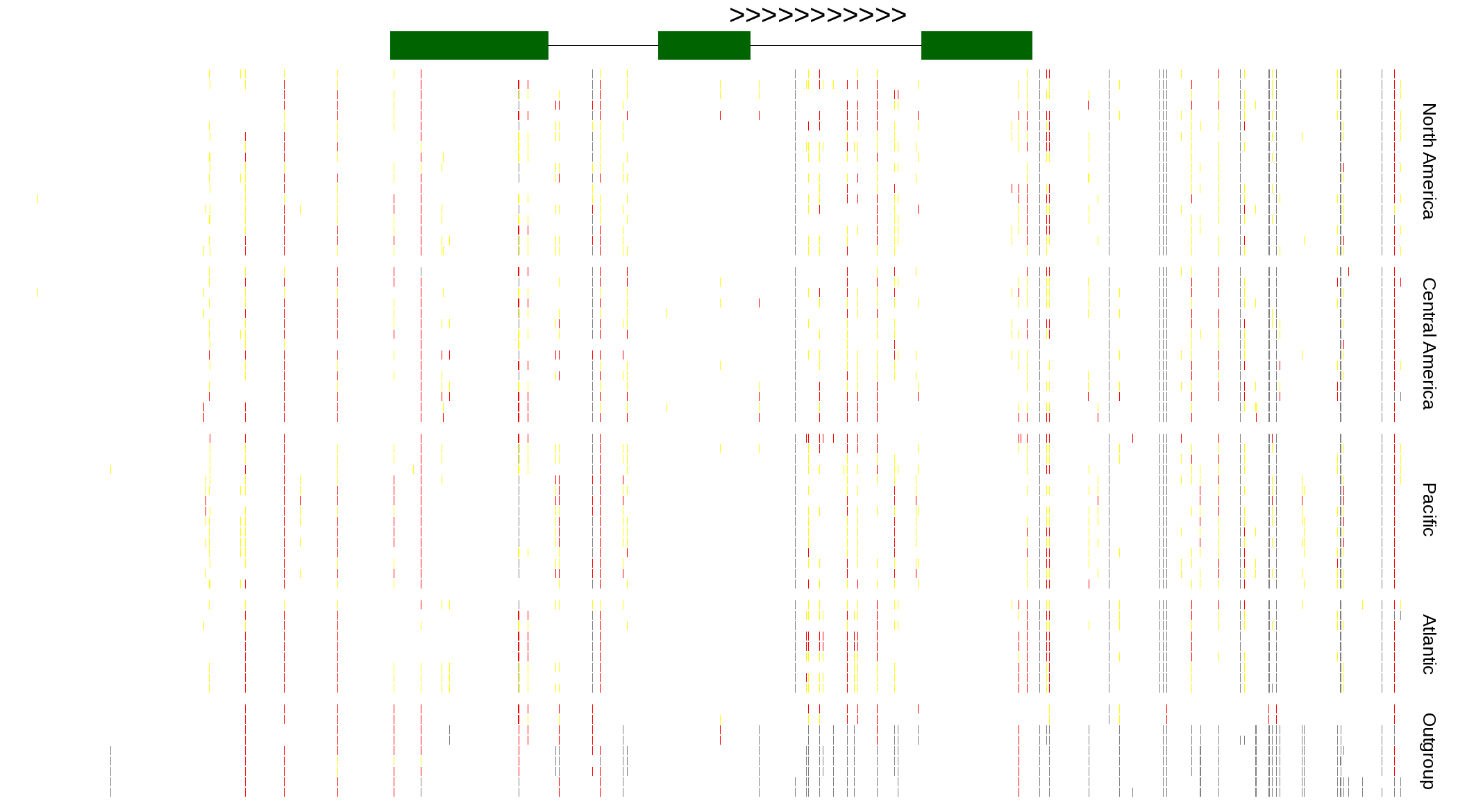
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS212228-TA
ATGGCTTTCCCAAGACACGTGGAGAATTTAAAGTTTGTTGTTAACCAAGGCATAACGCATCTGGTAACACTAACAGCAGGGAAGAAGCCTCCAGTGGACGGTATTGCCAGATTAAGATGGACTGAAATACCCATAGAAGAATTTGGAAATCCTACAGTCGAGCAAATAAAGACCTTTATCGATGTTTGTAAAAGATCAGATAAAAATGGCGAGGTGATCGGTATCCACTGCCGTCAAGGCCGCGGTCGTTCCGGGGTGATGTTGGCCAGCTATCTCGTACATTTCCACCGTTTCCTTCCTGACCAAGCAATGAACGTTATACGAATGATACGACCAGGTTCATTGGATTTTGAGGAACATGAAGAGGCAGTTGGCAAATATTTTGAATACTTGACTGAAAGCAATCCGTTGAGATTCGGAGTCAGTGGAGACGTTATGGAGGAGTTTATTGAAGCGGCCAAGGAAGCCACCAAAAATGCCTTGTAA
>DPOGS212228-PA
MAFPRHVENLKFVVNQGITHLVTLTAGKKPPVDGIARLRWTEIPIEEFGNPTVEQIKTFIDVCKRSDKNGEVIGIHCRQGRGRSGVMLASYLVHFHRFLPDQAMNVIRMIRPGSLDFEEHEEAVGKYFEYLTESNPLRFGVSGDVMEEFIEAAKEATKNAL-