| DPOGS212416 | ||
|---|---|---|
| Transcript | DPOGS212416-TA | 627 bp |
| Protein | DPOGS212416-PA | 208 aa |
| Genomic position | DPSCF300258 - 72133-77826 | |
| RNAseq coverage | 1771x (Rank: top 7%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL013249 | 3e-57 | 86.73% | |
| Bombyx | BGIBMGA014446-TA | 2e-34 | 95.45% | |
| Drosophila | ATPCL-PD | 2e-39 | 63.72% | |
| EBI UniRef50 | UniRef50_Q6AWP8 | 3e-37 | 63.72% | RE70805p n=33 Tax=cellular organisms RepID=Q6AWP8_DROME |
| NCBI RefSeq | XP_002081694.1 | 5e-38 | 63.72% | GD25577 [Drosophila simulans] |
| NCBI nr blastp | gi|195583780 | 8e-37 | 63.72% | GD25577 [Drosophila simulans] |
| NCBI nr blastx | gi|281363477 | 6e-36 | 63.72% | ATP citrate lyase, isoform F [Drosophila melanogaster] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005524 | 2.7e-13 | ATP binding | |
| GO:0016874 | 2.7e-13 | ligase activity | ||
| KEGG pathway | dsi:Dsim_GD25577 | 1e-37 | ||
| K01648 (ACLY) | maps-> | Citrate cycle (TCA cycle) | ||
| Reductive carboxylate cycle (CO2 fixation) | ||||
| InterPro domain | [82-158] IPR013816 | 2.7e-13 | ATP-grasp fold, subdomain 2 | |
| Orthology group | ||||
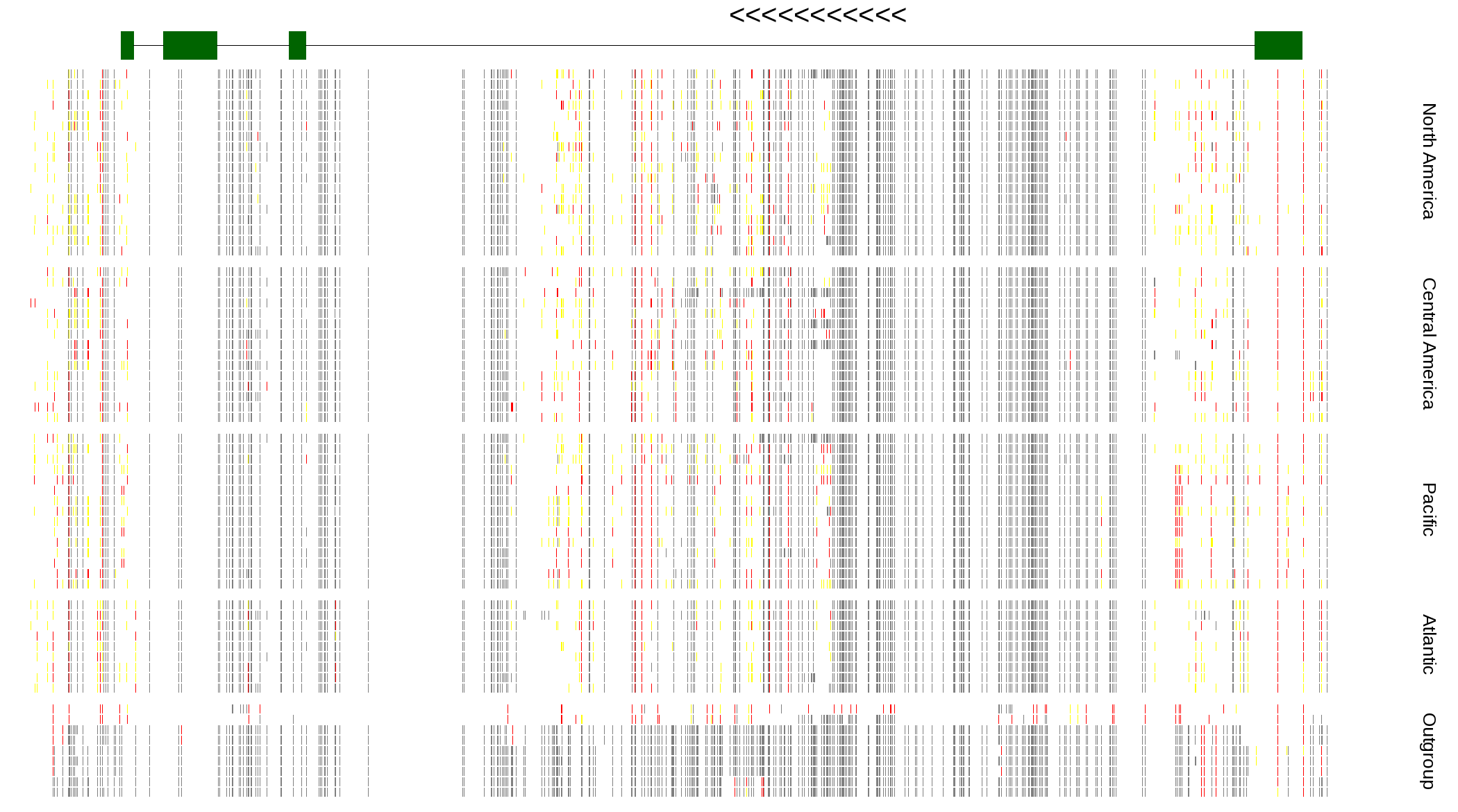
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS212416-TA
ATGGTGAAGTTGGGTTTGGCGATGGTGGTCGGCGTGGCGCCGGCGCTTGTCTCCTGGTGGAGGGGGCAGGGTCCGTGGCTGTCGCTTCCGTTGACCTTACAGAGCCCATGCTCAGAAAAGTCCATGAAATGGCCCCAGCGAAGATCACCTTCATTATCCGATTGCTTGGAATCCACCAGCGATGACGATTCCTCGCGGTCTATTGTATGCCACGTGAGGACTAAGGTCATCCCCGTGGACACTTTCCCATCCATCGAGGACATTGATCGTGTACTGTTGAAGAACATCAAAGCCACAACAACAAAGAGGATCCTGTCAAAGTTCATCGTGAGCCTGTACCGCGTGTTCGTGAACCTGTACTTCACGTACATGGAGATCAACCCGGTGGTGGTGACCAACGAGCGAGTCTACCTCCTTGACCTGGCCGCCAAGTTGGATCAGACGGCGGATTTCATATGCGCCAAGAACTGGGGGGAGATCACCTTCCCCCCGCCCTTCGGCAGGGACGCGTACCCCGAGGAAGCTCATATAGCTGATCTGGACGCTAAGAGCGGGGCTAGCTTGAAGGAAGTTGCCATATTTGCGAGTTGTGACGCTATCTTCAACCGCGGAACTATTTCCTCGTGA
>DPOGS212416-PA
MVKLGLAMVVGVAPALVSWWRGQGPWLSLPLTLQSPCSEKSMKWPQRRSPSLSDCLESTSDDDSSRSIVCHVRTKVIPVDTFPSIEDIDRVLLKNIKATTTKRILSKFIVSLYRVFVNLYFTYMEINPVVVTNERVYLLDLAAKLDQTADFICAKNWGEITFPPPFGRDAYPEEAHIADLDAKSGASLKEVAIFASCDAIFNRGTISS-