| DPOGS212991 | ||
|---|---|---|
| Transcript | DPOGS212991-TA | 525 bp |
| Protein | DPOGS212991-PA | 174 aa |
| Genomic position | DPSCF300024 - 865937-867212 | |
| RNAseq coverage | 506x (Rank: top 25%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL007697 | 7e-56 | 63.58% | |
| Bombyx | BGIBMGA006905-TA | 9e-54 | 58.72% | |
| Drosophila | Fuca-PB | 1e-30 | 39.49% | |
| EBI UniRef50 | UniRef50_D6WGK4 | 4e-37 | 47.02% | Putative uncharacterized protein n=3 Tax=Tribolium castaneum RepID=D6WGK4_TRICA |
| NCBI RefSeq | XP_971511.1 | 7e-38 | 47.02% | PREDICTED: similar to AGAP007285-PA [Tribolium castaneum] |
| NCBI nr blastp | gi|91078838 | 1e-36 | 47.02% | PREDICTED: similar to AGAP007285-PA [Tribolium castaneum] |
| NCBI nr blastx | gi|270003719 | 8e-38 | 47.02% | hypothetical protein TcasGA2_TC002989 [Tribolium castaneum] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005975 | 2.1e-45 | carbohydrate metabolic process | |
| GO:0004560 | 2.1e-45 | alpha-L-fucosidase activity | ||
| GO:0006004 | 4.5e-18 | fucose metabolic process | ||
| GO:0043169 | 3.6e-16 | cation binding | ||
| GO:0003824 | 3.6e-16 | catalytic activity | ||
| KEGG pathway | tca:660163 | 2e-37 | ||
| K01206 (E3.2.1.51, FUCA) | maps-> | Other glycan degradation | ||
| InterPro domain | [1-171] IPR000933 | 2.1e-45 | Glycoside hydrolase, family 29 | |
| [11-32] IPR016286 | 4.5e-18 | Glycoside hydrolase, family 29, bacteria/metazoa/fungi | ||
| [4-61] IPR017853 | 1.1e-17 | Glycoside hydrolase, superfamily | ||
| [4-61] IPR013781 | 3.6e-16 | Glycoside hydrolase, subgroup, catalytic core | ||
| Orthology group | ||||
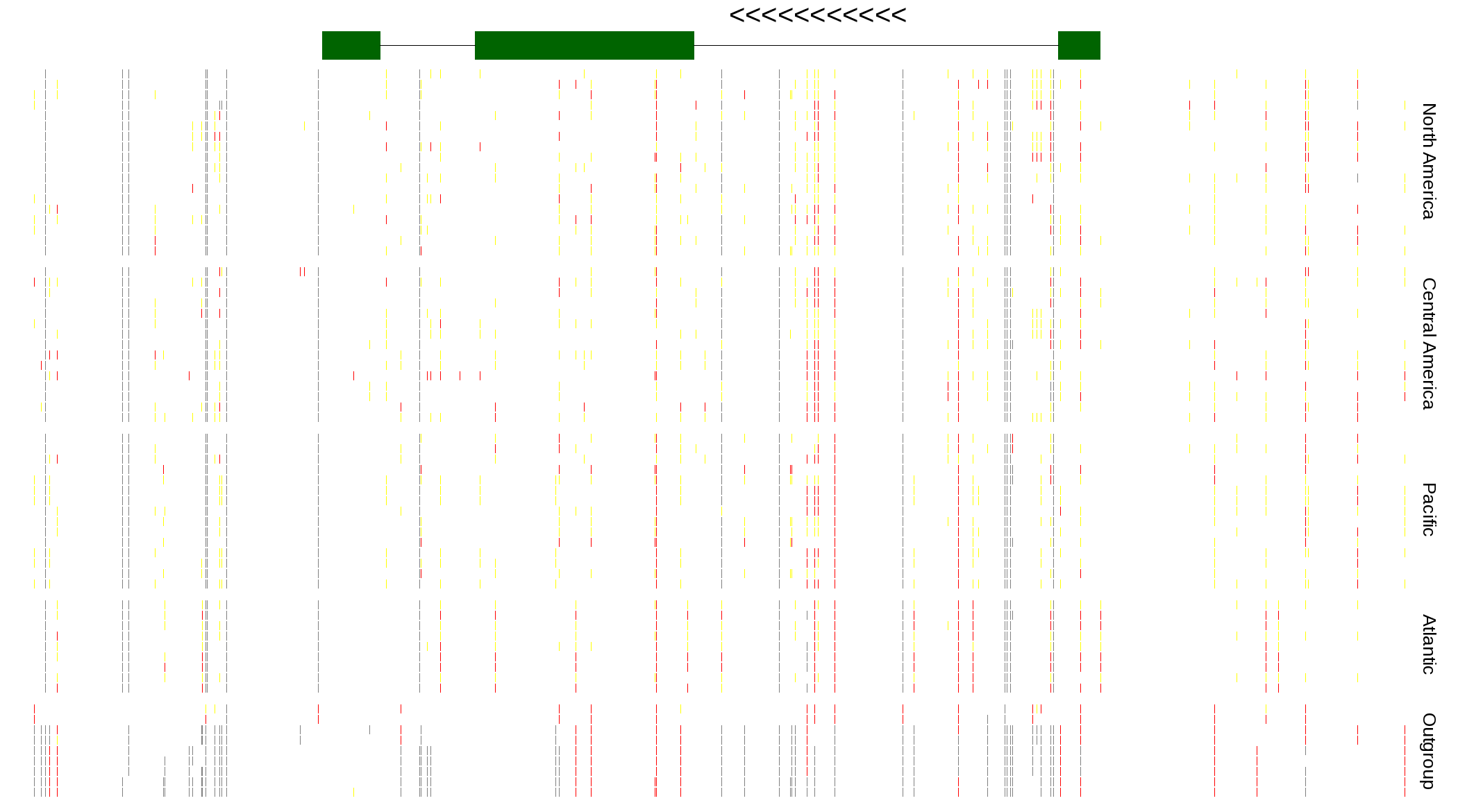
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS212991-TA
ATGGCTATAGAAGATGTTCTTAGTATTGAGCATTTGTTGAAAGAAGTAGTGTCTACTGTCAGCTGCGGAGGCAATGCTCTAATAAACGTCGGGCCAACGAAGGAAGGTACCATCGCTGCTATATTCCAAGAGCGATTGCTGGCTCTCGGCGACTGGTTGTCCTTGAACGGCGAAGCTATCTACGGCAGCTCGCCGTGGCATTCCCAGAACGACACCAGGAACGGCGACGTCTGGTACACCTGTGTCAAGAGCACCTTCAACGCCCGTCATCCGTCATCGAGGCCGAGGAGAGCTGATATGATAACAGCCATATACGCCGTTGTATTGAAATGGCCTGAAAACAACTTGCTGGTTATCAGTGACATTGTTCAGGATCTACACTCCGGTACTTATGAGGTCACTATGATCGGCTCCGAGGGGCACTTGAAGTGGAACATTGATAGTGGCCTCGTATATATAAGGCTTCCTGATAAGGCTACCGTCAAATCAAACTACGCCTGGACGCTCAAATTTAAACCAAAATAA
>DPOGS212991-PA
MAIEDVLSIEHLLKEVVSTVSCGGNALINVGPTKEGTIAAIFQERLLALGDWLSLNGEAIYGSSPWHSQNDTRNGDVWYTCVKSTFNARHPSSRPRRADMITAIYAVVLKWPENNLLVISDIVQDLHSGTYEVTMIGSEGHLKWNIDSGLVYIRLPDKATVKSNYAWTLKFKPK-