| DPOGS213239 | ||
|---|---|---|
| Transcript | DPOGS213239-TA | 369 bp |
| Protein | DPOGS213239-PA | 122 aa |
| Genomic position | DPSCF300124 - 389993-390553 | |
| RNAseq coverage | 214x (Rank: top 46%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL004705 | 2e-62 | 94.87% | |
| Bombyx | BGIBMGA009518-TA | 2e-51 | 76.42% | |
| Drosophila | CG5731-PA | 2e-40 | 65.18% | |
| EBI UniRef50 | UniRef50_E0VBI5 | 1e-41 | 69.44% | Alpha-N-acetylgalactosaminidase, putative n=5 Tax=Bilateria RepID=E0VBI5_PEDHC |
| NCBI RefSeq | XP_001606799.1 | 3e-44 | 83.51% | PREDICTED: similar to ENSANGP00000020847 [Nasonia vitripennis] |
| NCBI nr blastp | gi|332025551 | 2e-49 | 88.30% | Alpha-N-acetylgalactosaminidase [Acromyrmex echinatior] |
| NCBI nr blastx | gi|322799781 | 3e-47 | 77.06% | hypothetical protein SINV_01689 [Solenopsis invicta] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008152 | 3.2e-31 | metabolic process | |
| GO:0003824 | 3.2e-31 | catalytic activity | ||
| GO:0004553 | 1.3e-11 | hydrolase activity, hydrolyzing O-glycosyl compounds | ||
| GO:0005975 | 1.3e-11 | carbohydrate metabolic process | ||
| KEGG pathway | ame:725899 | 5e-41 | ||
| K01204 (NAGA) | maps-> | Lysosome | ||
| Glycosphingolipid biosynthesis - globo series | ||||
| InterPro domain | [24-117] IPR013785 | 3.2e-31 | Aldolase-type TIM barrel | |
| [24-116] IPR017853 | 2.1e-30 | Glycoside hydrolase, superfamily | ||
| [26-45] IPR002241 | 1.3e-11 | Glycoside hydrolase, family 27 | ||
| [31-118] IPR000111 | 2.1e-07 | Glycoside hydrolase, clan GH-D | ||
| Orthology group | MCL25617 | Lepidoptera specific | ||
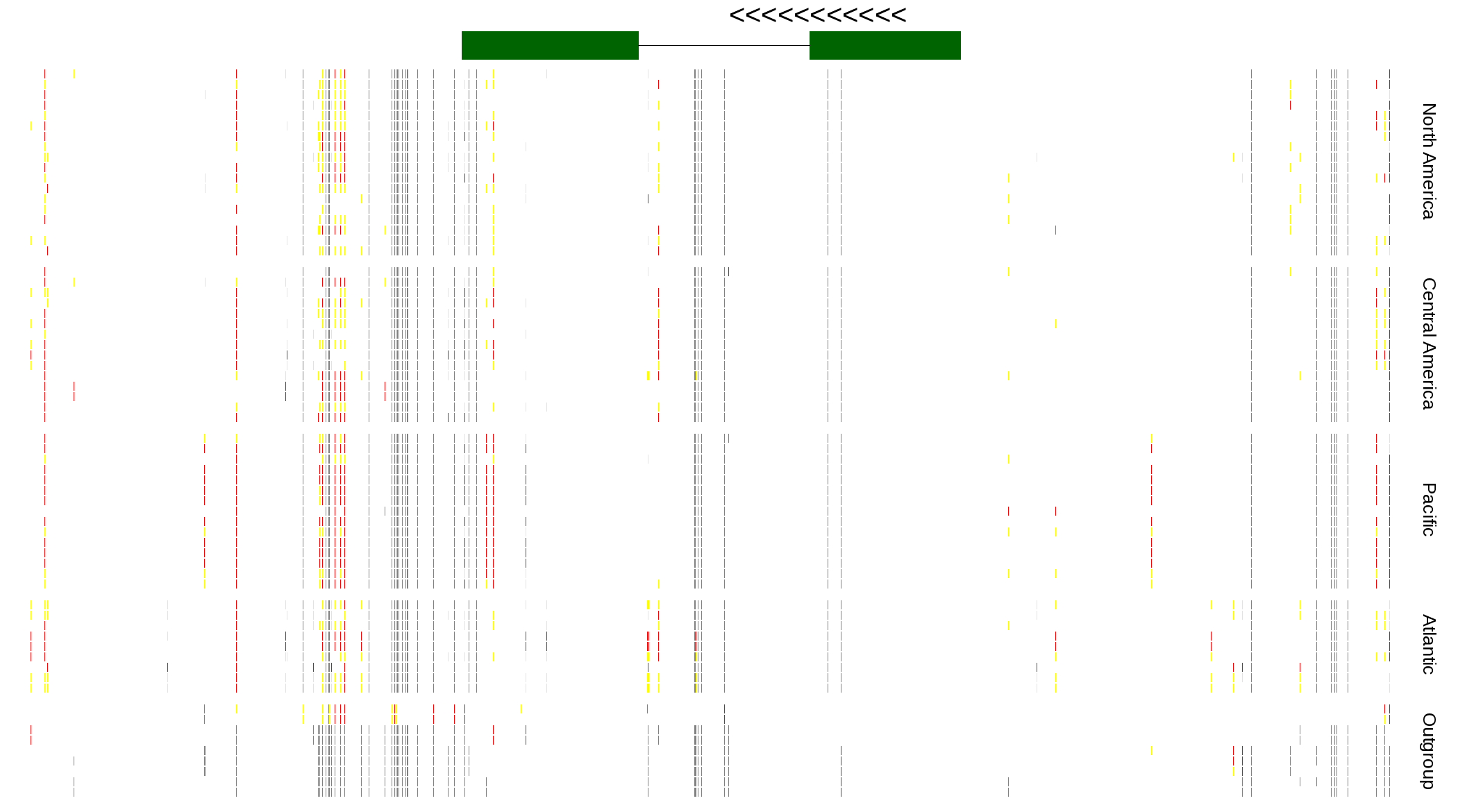
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS213239-TA
ATGGGGTCATGGAAACTGTGCGTGTGGATATTGTTTGTGGTTGTGTGTATCCTTGGGAAGATATCAGGTCTTGATAATGGGTTGGCATTGACCCCTCCCATGGGATGGCTGGCTTGGGAGAGGTTCAGATGTAACACGGACTGTAAAAATGATCCTGATAATTGTATAAGCGATCGTCTGTTCAGAACGATGACGGATATTCTGGTAGCCGAGGGTTATGCTGCAGCTGGTTACGAATACGTGAACGTGGACGACTGCTGGCCTGAAAGAGAGAGAGATCCGAGGGGGAAGCTTGTACCTGACAGAGAACGCTTCCCTTATGGAATGAAGAGCCTGTCTGATTACGTGAGTAATATATATATAAATTAA
>DPOGS213239-PA
MGSWKLCVWILFVVVCILGKISGLDNGLALTPPMGWLAWERFRCNTDCKNDPDNCISDRLFRTMTDILVAEGYAAAGYEYVNVDDCWPERERDPRGKLVPDRERFPYGMKSLSDYVSNIYIN-