| DPOGS213334 | ||
|---|---|---|
| Transcript | DPOGS213334-TA | 276 bp |
| Protein | DPOGS213334-PA | 91 aa |
| Genomic position | DPSCF300563 - 26656-27195 | |
| RNAseq coverage | 0x (Rank: top 99%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL016675 | 1e-32 | 65.56% | |
| Bombyx | BGIBMGA004721-TA | 4e-20 | 46.88% | |
| Drosophila | Cyp4c3-PA | 6e-21 | 42.55% | |
| EBI UniRef50 | UniRef50_D2JLJ9 | 2e-27 | 59.55% | Cytochrome P450 CYP405A3 n=3 Tax=Ditrysia RepID=D2JLJ9_9NEOP |
| NCBI RefSeq | XP_972577.1 | 5e-21 | 53.26% | PREDICTED: similar to pheromone-degrading enzyme [Tribolium castaneum] |
| NCBI nr blastp | gi|308316602 | 2e-30 | 69.66% | cytochrome P450 CYP405A2 [Zygaena filipendulae] |
| NCBI nr blastx | gi|308316602 | 1e-29 | 69.66% | cytochrome P450 CYP405A2 [Zygaena filipendulae] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0009055 | 7.6e-30 | electron carrier activity | |
| GO:0020037 | 7.6e-30 | heme binding | ||
| GO:0016705 | 7.6e-30 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | ||
| GO:0005506 | 7.6e-30 | iron ion binding | ||
| GO:0055114 | 7.6e-30 | oxidation-reduction process | ||
| KEGG pathway | mmu:230639 | 1e-19 | ||
| K07425 (CYP4A) | maps-> | Arachidonic acid metabolism | ||
| Fatty acid metabolism | ||||
| Vascular smooth muscle contraction | ||||
| PPAR signaling pathway | ||||
| Retinol metabolism | ||||
| InterPro domain | [1-90] IPR001128 | 7.6e-30 | Cytochrome P450 | |
| [46-70] IPR002401 | 3.6e-08 | Cytochrome P450, E-class, group I | ||
| Orthology group | ||||
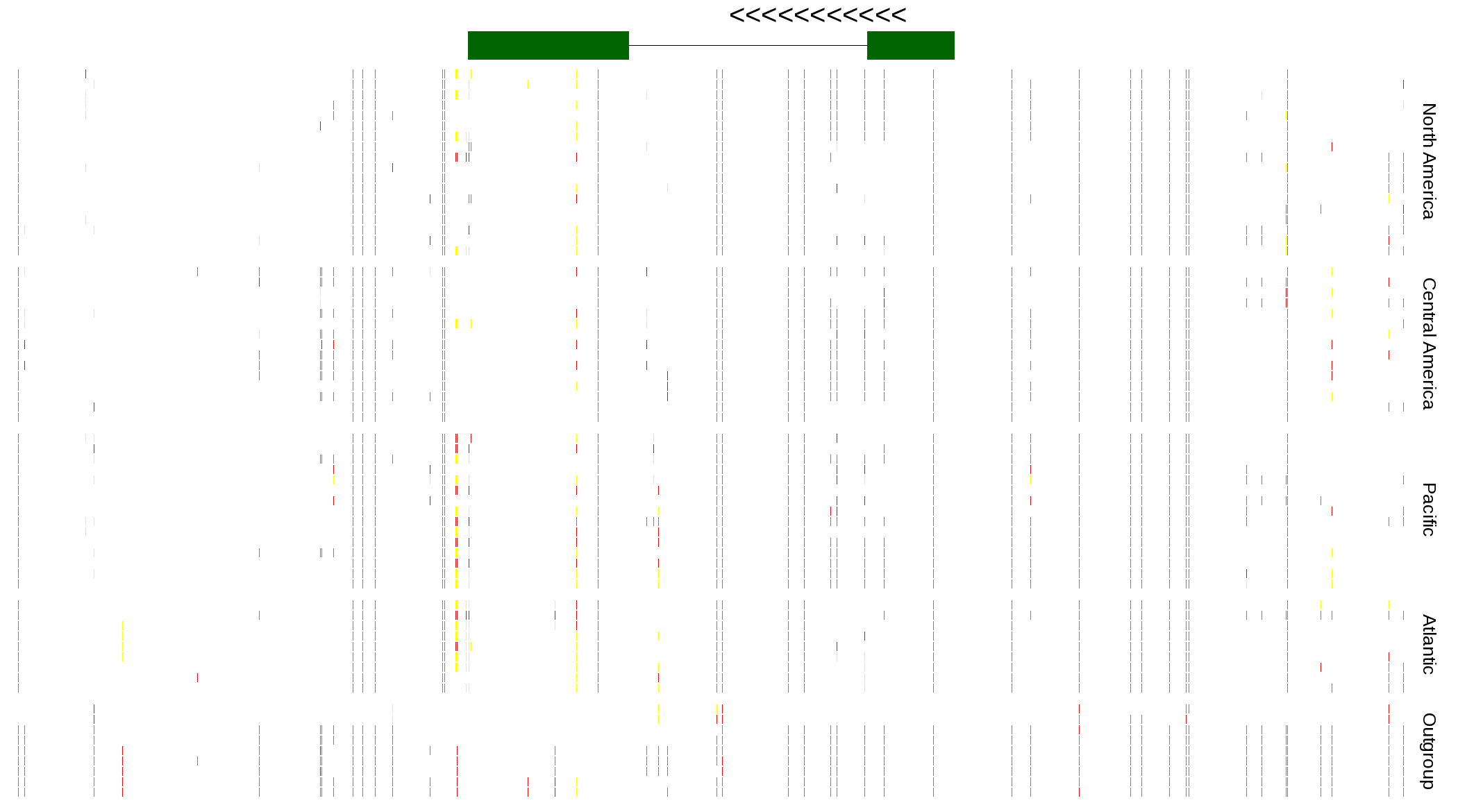
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS213334-TA
ATGAAGTACCTTGAGGCTGTATTAAAAGAAAGCGTTCGAGTAATACCAACTGTGACTAAGATCGGAAGACAATTACACGAGGATCTTAAATTTAAAGATGGACGCATAGCTCCGGCAGGAAGTTCAGTTGTTGTTTTCTTCGAAGCAATGTATCAAAATCCCAAAATATTTCCTGAACCGGAAAAATATGATCCTGAAAGGTTTTTTAACAATATGCATACTTTTGCTTTCGTTCCATTTAGTGCAGGGCCAAGAAATTGTATAGGTAAGTATTAA
>DPOGS213334-PA
MKYLEAVLKESVRVIPTVTKIGRQLHEDLKFKDGRIAPAGSSVVVFFEAMYQNPKIFPEPEKYDPERFFNNMHTFAFVPFSAGPRNCIGKY-