| DPOGS213773 | ||
|---|---|---|
| Transcript | DPOGS213773-TA | 729 bp |
| Protein | DPOGS213773-PA | 242 aa |
| Genomic position | DPSCF300212 + 255557-256285 | |
| RNAseq coverage | 0x (Rank: top 99%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL010466 | 1e-53 | 41.18% | |
| Bombyx | BGIBMGA009243-TA | 2e-50 | 37.39% | |
| Drosophila | FucTC-PB | 2e-22 | 31.28% | |
| EBI UniRef50 | UniRef50_F4X2Z6 | 1e-24 | 32.64% | Alpha-(1,3)-fucosyltransferase C n=1 Tax=Acromyrmex echinatior RepID=F4X2Z6_ACREC |
| NCBI RefSeq | XP_001120699.1 | 2e-24 | 32.02% | PREDICTED: similar to Alpha-(1,3)-fucosyltransferase C (Galactoside 3-L-fucosyltransferase) [Apis mellifera] |
| NCBI nr blastp | gi|332018601 | 3e-24 | 32.64% | Alpha-(1,3)-fucosyltransferase C [Acromyrmex echinatior] |
| NCBI nr blastx | gi|332018601 | 3e-26 | 32.13% | Alpha-(1,3)-fucosyltransferase C [Acromyrmex echinatior] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0016020 | 4.1e-31 | membrane | |
| GO:0008417 | 4.1e-31 | fucosyltransferase activity | ||
| GO:0006486 | 4.1e-31 | protein glycosylation | ||
| KEGG pathway | api:100161726 | 9e-16 | ||
| K00753 (E2.4.1.214) | maps-> | N-Glycan biosynthesis | ||
| InterPro domain | [19-241] IPR001503 | 4.1e-31 | Glycosyl transferase, family 10 | |
| Orthology group | ||||
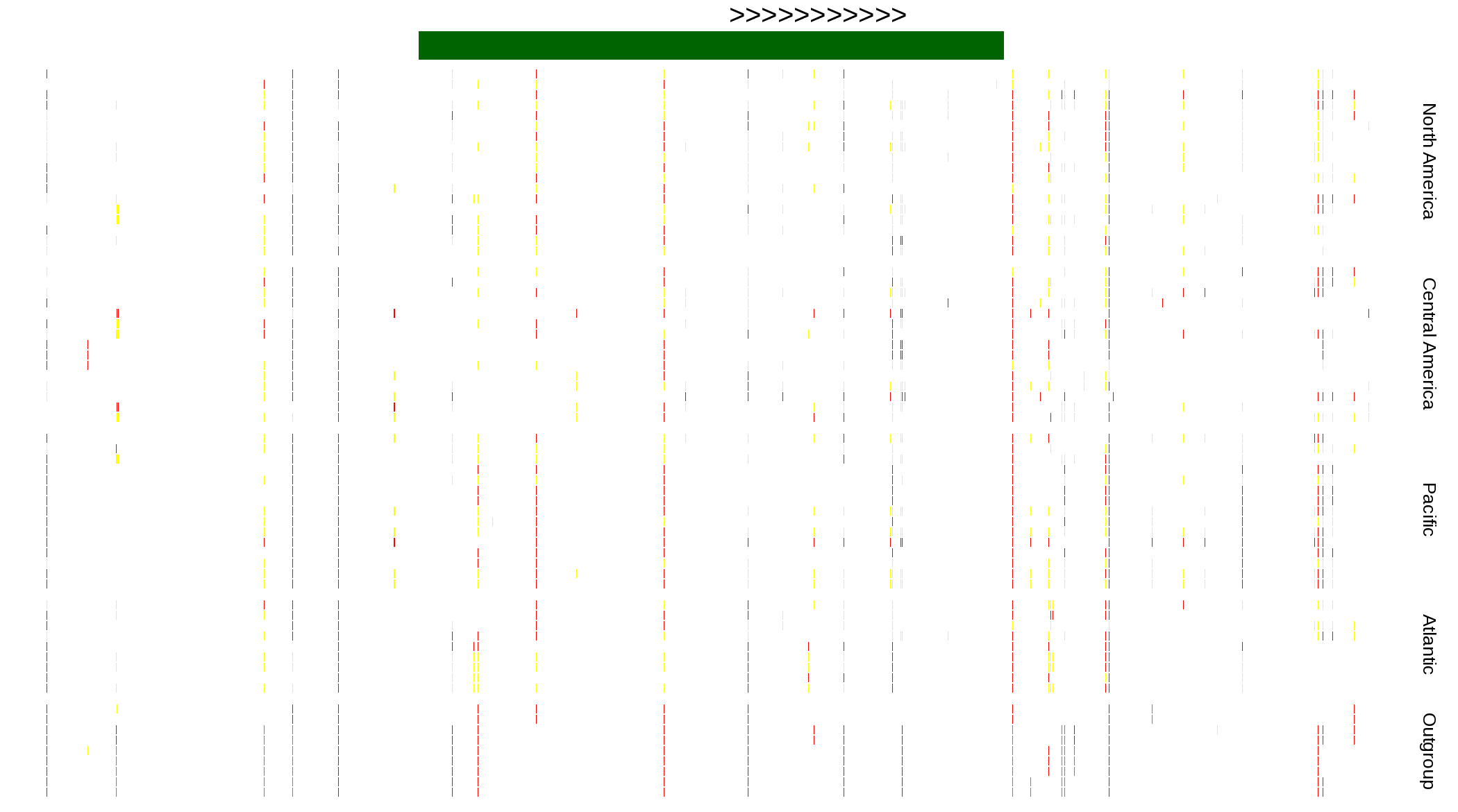
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS213773-TA
ATGTTTAATGATACAGAAAATAAAATTGGCGAAGGACAAAAAGCTTTTATCGATCGTGATTGTAGATACGTGAACTGTTATTTAACGACTAAGAAGGATTTCCTTAACAATGACGTCACCAATTTTAATGCAATAGTATTTGACATAAACAAGATCAAGATGTGGAAAAAGATGTTCTTTCCAAAGCTGAGGTCTTATGAGCAGAAATATATTTTCTACAGTGACGTTTCCTCTGACGATGTGCCTATATGTAATATCAATATGGATAATTATTTTAATTGGACTTGGACGTACAAAATTAATTCTGATATTGTAAGCCCTTTTATTGAAGTTAAAGACTTAAAAGGTAATGTTGTTGCTCCCAGACCAGTGGTGAATTGGAATTCTAATATGACAGTACTAGATGAAGATGAAATAAAGCATTTGAAGCAAAAGAAAAAGGCAATGGCTTGGGTAGTTACAAAATGTCACACTAGAAACAATAGATTATTGCTGGCTAGAAGATTGAGACGGGGTTTTGAACAGAACGATTTAATATTTGATATATATGGATGTGGTCATAAGAATTGTCCTAAAGGAGGATGTATGAAAGCAATTGAAAGAGAGTATTACTTCTACTTTGTCGCTGAAGCCAGCTTTGATGAAGATTATGTAACTGATGAGGTCCTGGCAGCCTACCATCATTATGCCGTTCCTGTTGTTTTGGGTGGAGCTAATTATCGCAGGTAA
>DPOGS213773-PA
MFNDTENKIGEGQKAFIDRDCRYVNCYLTTKKDFLNNDVTNFNAIVFDINKIKMWKKMFFPKLRSYEQKYIFYSDVSSDDVPICNINMDNYFNWTWTYKINSDIVSPFIEVKDLKGNVVAPRPVVNWNSNMTVLDEDEIKHLKQKKKAMAWVVTKCHTRNNRLLLARRLRRGFEQNDLIFDIYGCGHKNCPKGGCMKAIEREYYFYFVAEASFDEDYVTDEVLAAYHHYAVPVVLGGANYRR-