| DPOGS214843 | ||
|---|---|---|
| Transcript | DPOGS214843-TA | 498 bp |
| Protein | DPOGS214843-PA | 165 aa |
| Genomic position | DPSCF300091 - 280712-321506 | |
| RNAseq coverage | 34x (Rank: top 74%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL007610 | 2e-31 | 95.59% | |
| Bombyx | BGIBMGA010012-TA | 7e-20 | 73.13% | |
| Drosophila | ss-PD | 3e-39 | 96.30% | |
| EBI UniRef50 | UniRef50_B4KDN6 | 2e-37 | 96.30% | GI23647 n=1 Tax=Drosophila mojavensis RepID=B4KDN6_DROMO |
| NCBI RefSeq | XP_394737.3 | 1e-38 | 97.53% | PREDICTED: similar to spineless CG6993-PA [Apis mellifera] |
| NCBI nr blastp | gi|345486455 | 2e-37 | 97.53% | PREDICTED: hypothetical protein LOC100123827 [Nasonia vitripennis] |
| NCBI nr blastx | gi|328780897 | 5e-37 | 75.89% | PREDICTED: LOW QUALITY PROTEIN: hypothetical protein LOC411264 [Apis mellifera] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005634 | 3.7e-07 | nucleus | |
| GO:0006355 | 3.7e-07 | regulation of transcription, DNA-dependent | ||
| KEGG pathway | ||||
| InterPro domain | [95-154] IPR011598 | 3.7e-07 | Helix-loop-helix DNA-binding | |
| [102-156] IPR001092 | 5e-06 | Helix-loop-helix DNA-binding domain | ||
| Orthology group | MCL18890 | Patchy | ||
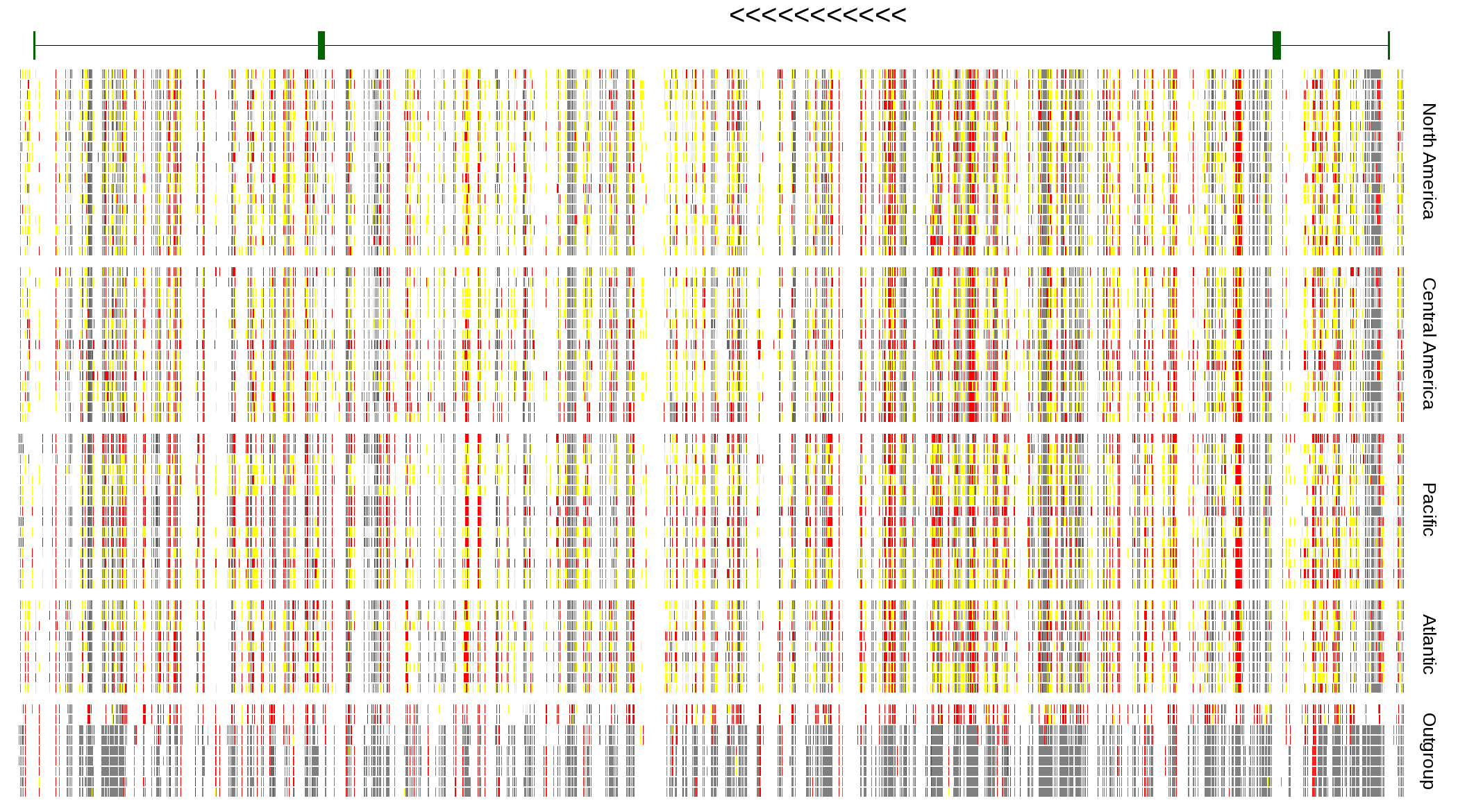
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS214843-TA
ATGCACCGTGGCTTGATTGACAGTTCCCTGCGAACATCCGTCAAGCTGTCGCCGCACATCCAGCCGAATTACCAGCCGTTTCACCCCGAGGACGGCTCAGCCGGCAAGATAGCCAGCGAGTCGAAGCTGCAACTGTGCGGCTGGGGCGGGCAGGGCTACGCTTTGGACGGCGTGGACACGAGCCTGCTTCGGCCGCCGCCGCAGCCTCACGCCAGCCAGCTAGGAACCGTTTACGCTACCAAGAGAAGGAGGAGGAATGGAAAAAGCATCAAACCACCTTCAAAAGACGGCGTCACGAAGAGCAATCCAAGTAAAAGACATCGAGAGCGTCTCAATGCGGAACTTGATACATTGGCGTCGCTGCTGCCCTTCGAGCAGAACATACTCAGCAAACTGGACCGACTCTCCATACTGCGACTATCAGTTAGCTACTTGCGGACCAAGAGCTACTTCCAAGAAGAAAGCTGTACGAGCCCGCTCATCCCAGCTGGGCAGTAA
>DPOGS214843-PA
MHRGLIDSSLRTSVKLSPHIQPNYQPFHPEDGSAGKIASESKLQLCGWGGQGYALDGVDTSLLRPPPQPHASQLGTVYATKRRRRNGKSIKPPSKDGVTKSNPSKRHRERLNAELDTLASLLPFEQNILSKLDRLSILRLSVSYLRTKSYFQEESCTSPLIPAGQ-