| DPOGS215804 | ||
|---|---|---|
| Transcript | DPOGS215804-TA | 579 bp |
| Protein | DPOGS215804-PA | 192 aa |
| Genomic position | DPSCF300041 + 2285895-2286473 | |
| RNAseq coverage | 968x (Rank: top 13%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL005814 | 5e-102 | 88.54% | |
| Bombyx | BGIBMGA003522-TA | 3e-97 | 87.70% | |
| Drosophila | Cyp1-PA | 5e-72 | 75.62% | |
| EBI UniRef50 | UniRef50_UPI000194C79A | 6e-71 | 76.25% | UPI000194C79A related cluster n=1 Tax=unknown RepID=UPI000194C79A |
| NCBI RefSeq | NP_001040172.1 | 2e-98 | 87.50% | peptidyl-prolyl cis-trans isomerase [Bombyx mori] |
| NCBI nr blastp | gi|114051666 | 4e-97 | 87.50% | peptidyl-prolyl cis-trans isomerase [Bombyx mori] |
| NCBI nr blastx | gi|114051666 | 3e-95 | 87.50% | peptidyl-prolyl cis-trans isomerase [Bombyx mori] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0006457 | 3.8e-78 | protein folding | |
| GO:0003755 | 3.8e-78 | peptidyl-prolyl cis-trans isomerase activity | ||
| KEGG pathway | ||||
| InterPro domain | [26-190] IPR002130 | 3.8e-78 | Peptidyl-prolyl cis-trans isomerase, cyclophilin-type | |
| [28-191] IPR015891 | 2.2e-75 | Cyclophilin-like | ||
| Orthology group | MCL24917 | Lepidoptera specific | ||
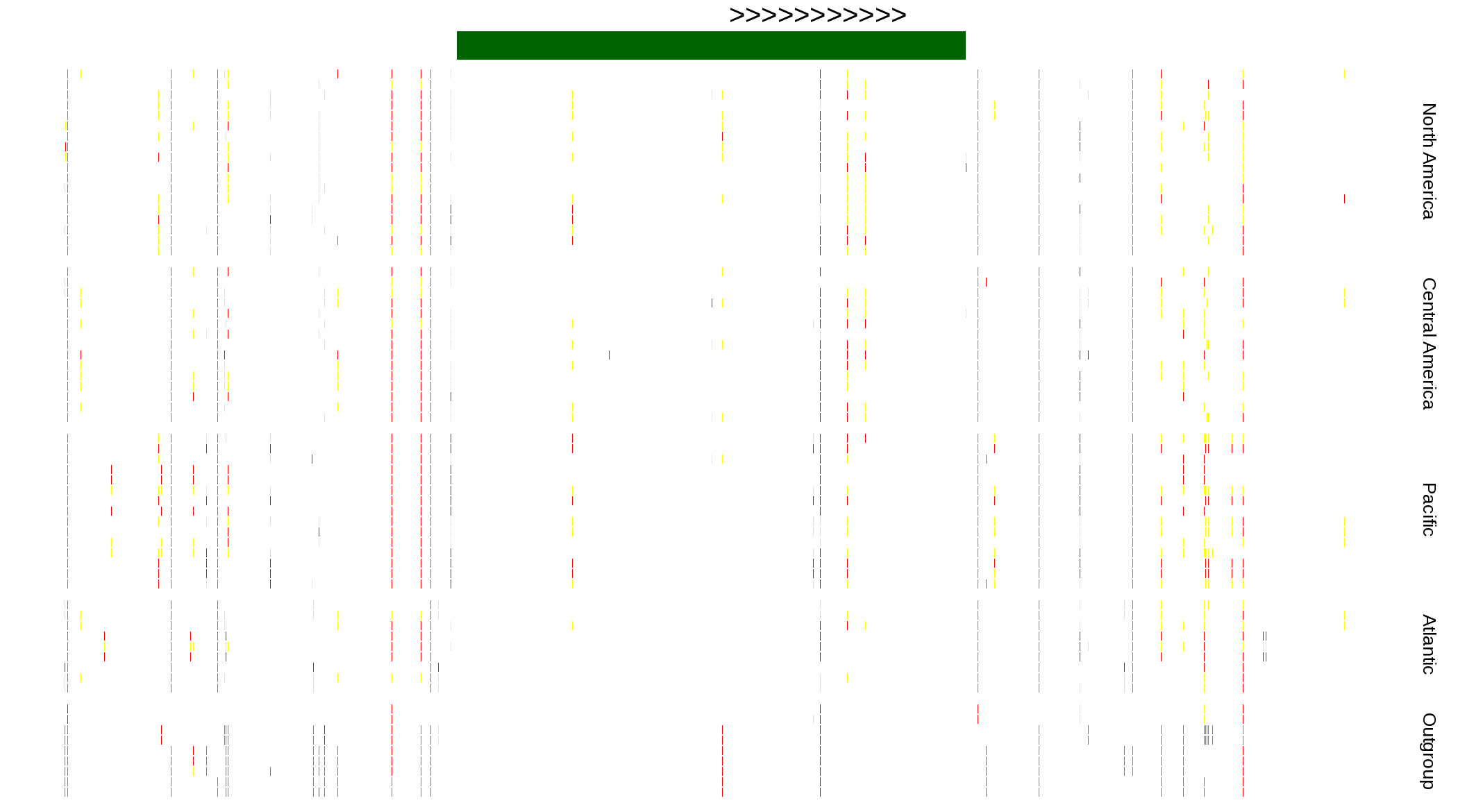
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS215804-TA
ATGGCTTTGACGGCAATGTTACGCCGTACCCTTATAAATGGCACATTATATTCAGCGGCCTCTTTGCGTTTTGCGTCTACACAGCAAGAAAAAAAAGTGTTCTTTGATGTCTCTGCTGATGGCGAACCCTTAGGCCGCATAGTTATGAAACTGAACACAGAAGAGGTCCCGAAGACTGCGGAGAATTTCAGAGCTCTGTGTACTGGAGAGAAAGGATTTGGTTATAAAGGTTCATCGTTCCACAGAATTATTCCTGACTTTATGTGTCAAGGAGGAGATTTCACTAATCACAATGGCACTGGTGGCAAGTCTATCTATGGTAGAACTTTCCAAGATGAGAACTTTAAGTTGAAGCACACTGGCCCCGGCATTCTATCTATGGCTAATGCTGGTCCTAACACAAACGGTTCTCAATTCTTCATTACAACCACTACAACCCCATGGCTGGATAGGAAACATGTTGTATTTGGATCAGTAGTTGAAGGAATGGATGTTGTGCGTAAAATGGAAAATCTTGGGTCAAAAAGTGGTAGACCTTCTAAAAAGGTTGTTATCCACGAATGTGGTGAGTTAGACTGA
>DPOGS215804-PA
MALTAMLRRTLINGTLYSAASLRFASTQQEKKVFFDVSADGEPLGRIVMKLNTEEVPKTAENFRALCTGEKGFGYKGSSFHRIIPDFMCQGGDFTNHNGTGGKSIYGRTFQDENFKLKHTGPGILSMANAGPNTNGSQFFITTTTTPWLDRKHVVFGSVVEGMDVVRKMENLGSKSGRPSKKVVIHECGELD-