| DPOGS201300 | ||
|---|---|---|
| Transcript | DPOGS201300-TA | 558 bp |
| Protein | DPOGS201300-PA | 185 aa |
| Genomic position | DPSCF300176 - 291805-294787 | |
| RNAseq coverage | 1707x (Rank: top 7%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL017251 | 2e-99 | 92.43% | |
| Bombyx | BGIBMGA003048-TA | 2e-91 | 85.87% | |
| Drosophila | Spase25-PA | 2e-50 | 49.18% | |
| EBI UniRef50 | UniRef50_G6D3W7 | 2e-103 | 100.00% | Signal peptidase complex subunit 2 n=2 Tax=Arthropoda RepID=G6D3W7_DANPL |
| NCBI RefSeq | NP_001040214.1 | 2e-91 | 86.96% | signal peptidase complex subunit 2 [Bombyx mori] |
| NCBI nr blastp | gi|114052092 | 4e-90 | 86.96% | signal peptidase complex subunit 2 [Bombyx mori] |
| NCBI nr blastx | gi|114052092 | 1e-87 | 86.96% | signal peptidase complex subunit 2 [Bombyx mori] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008233 | 3.7e-48 | peptidase activity | |
| GO:0016021 | 3.7e-48 | integral to membrane | ||
| GO:0006465 | 3.7e-48 | signal peptide processing | ||
| GO:0005787 | 3.7e-48 | signal peptidase complex | ||
| KEGG pathway | ame:410218 | 1e-52 | ||
| K12947 (SPCS2, SPC2) | maps-> | Protein export | ||
| InterPro domain | [19-177] IPR009582 | 3.7e-48 | Signal peptidase complex subunit 2 | |
| Orthology group | MCL14824 | Single-copy universal gene | ||
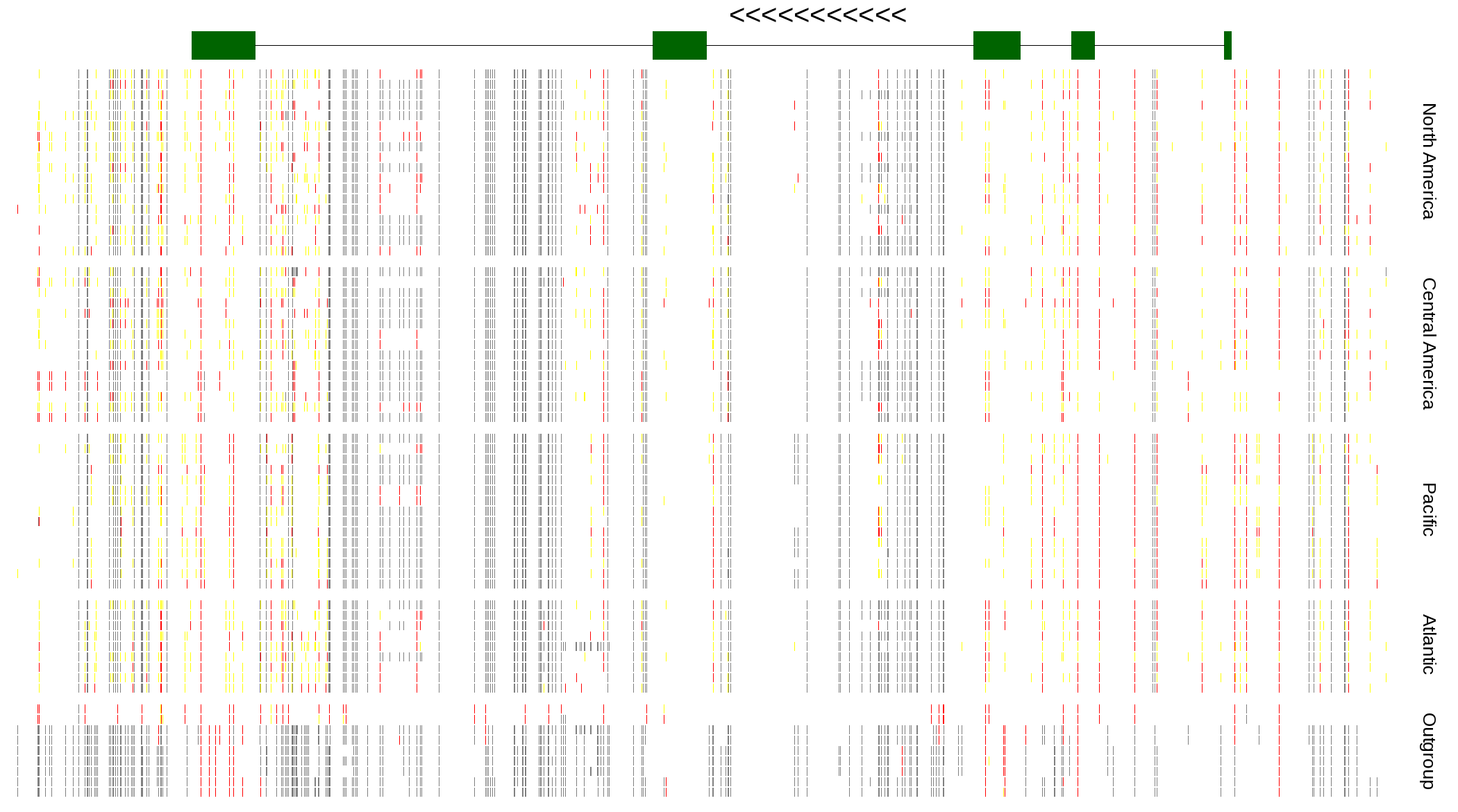
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS201300-TA
ATGGTGAACGAAACCCCAGAGGTTGTTAAAATCAACAAATGGGATGGAGCCGCTGCTAAAAATGCTATTGATGATGCAATCAGAGAGGTGTTTACCGGAGAGCTGAAATGTAAGGAGAGTTTTGCTTTGATCGACGGACGGTTATTTCTATGTGCCTTAGCTGTAGGAGTGGCCCTGTATGCTTTGCTCTGGGACTATCTCTATCCATTCCCACAGTCCAAACTCGTCCTTATCATCTGTGTATCCTCATATTTTATTTTAATGAGCATCCTTACTTTATACACTACATTCAAAGAAAAAGGCATTTTTGTTGTGGCCAAAGAGAAAACGGGAAACAACTCGAGAGTCTGGGAAGCAAGCTCTTATGTTAAAAAGCACGATGACAAATATAGTTTAGTTCTAGTGATGCGCGATGCTAACGGCAAGACCCGGGAAGCAACTGTAAACAAGTCCTTTGCTAATTACATTGATAACAACGGAACAGTTGTCCAGAGCATCATCATCAACGAAATAACGAAGCTCTACAACTCTTTGTCTTCGGAGAAGAAAGAAAAGTGA
>DPOGS201300-PA
MVNETPEVVKINKWDGAAAKNAIDDAIREVFTGELKCKESFALIDGRLFLCALAVGVALYALLWDYLYPFPQSKLVLIICVSSYFILMSILTLYTTFKEKGIFVVAKEKTGNNSRVWEASSYVKKHDDKYSLVLVMRDANGKTREATVNKSFANYIDNNGTVVQSIIINEITKLYNSLSSEKKEK-