| DPOGS202818 | ||
|---|---|---|
| Transcript | DPOGS202818-TA | 528 bp |
| Protein | DPOGS202818-PA | 175 aa |
| Genomic position | DPSCF300018 + 440608-443019 | |
| RNAseq coverage | 85x (Rank: top 63%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL003642 | 3e-65 | 96.03% | |
| Bombyx | BGIBMGA006995-TA | 1e-36 | 55.24% | |
| Drosophila | chm-PA | 2e-54 | 70.63% | |
| EBI UniRef50 | UniRef50_D6X346 | 4e-56 | 71.53% | Putative uncharacterized protein n=2 Tax=Tribolium castaneum RepID=D6X346_TRICA |
| NCBI RefSeq | XP_970403.2 | 6e-57 | 71.53% | PREDICTED: similar to chameau CG5229-PA [Tribolium castaneum] |
| NCBI nr blastp | gi|189241087 | 1e-55 | 71.53% | PREDICTED: similar to chameau CG5229-PA [Tribolium castaneum] |
| NCBI nr blastx | gi|270013354 | 2e-54 | 71.53% | hypothetical protein TcasGA2_TC011945 [Tribolium castaneum] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005634 | 6.7e-40 | nucleus | |
| GO:0006355 | 6.7e-40 | regulation of transcription, DNA-dependent | ||
| GO:0016747 | 6.7e-40 | transferase activity, transferring acyl groups other than amino-acyl groups | ||
| KEGG pathway | ||||
| InterPro domain | [32-170] IPR016181 | 7.8e-50 | Acyl-CoA N-acyltransferase | |
| [32-143] IPR002717 | 6.7e-40 | MOZ/SAS-like protein | ||
| Orthology group | MCL20685 | Insect specific | ||
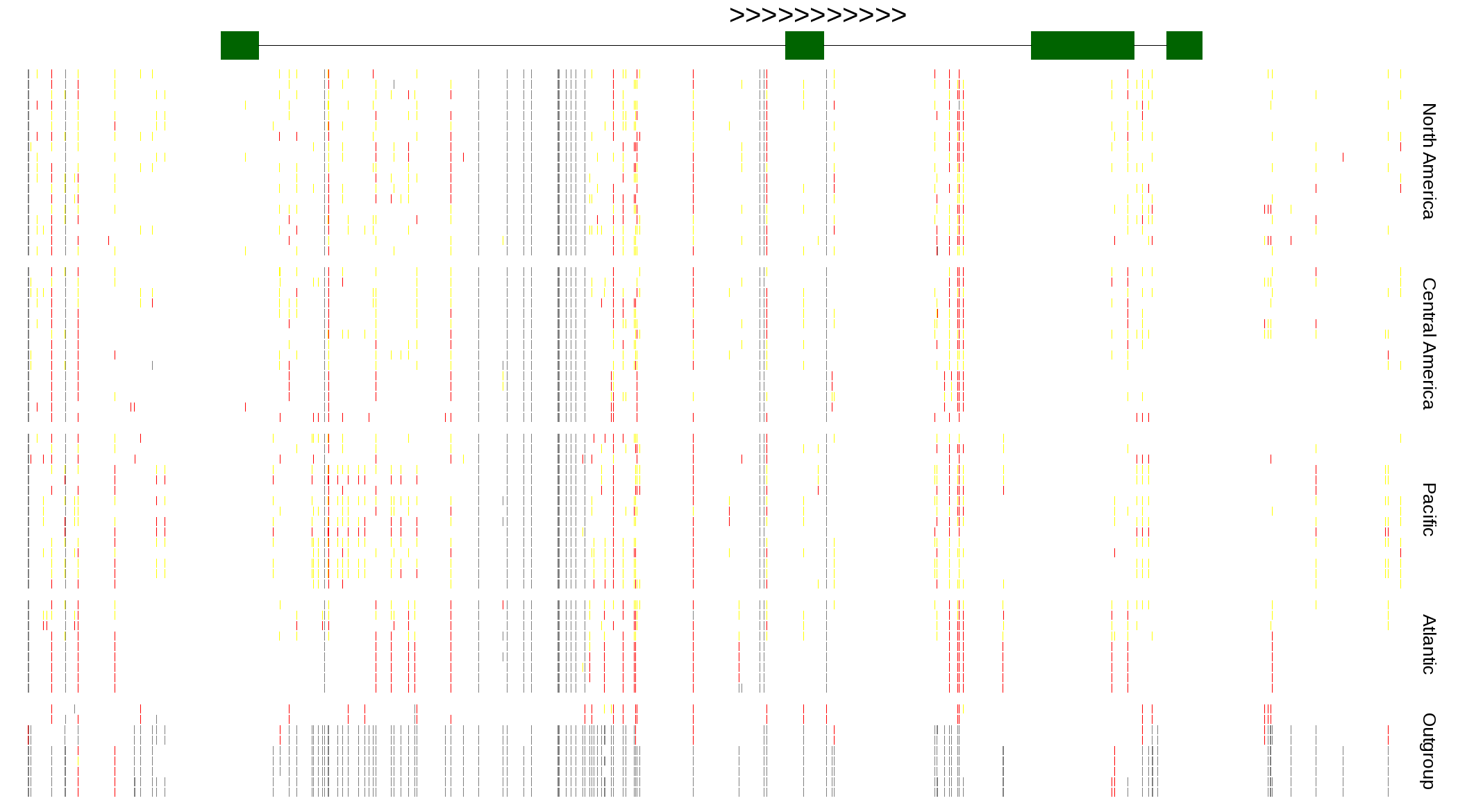
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS202818-TA
ATGATAAGTCTGGAATGGAATCCGTACACAGACGTCGAGACAGCGGGGTCGGGGTCGACGAACCCAGGGCTGCCACCGGTTATAGATTGGGAGGAGAAAAACTCGTTCTTGAACTACAACGTGTCTTGTATACTGACCCTACCGCCCTACCAGCGTCAGGGCTACGGCAGGCTGCTCATCGATTTCAGTTACCTCTTAACTAAAGTGGAGGGTAAAGTGGGTTCGCCGGAGACCCCGCTGTCGGACCTGGGGCTCATATCGTACAGGTCATACTGGAAGGAGGCTCTATTGAAGAGGCTCTGTTCGGCGTCAGGGTCGACTCTATGCATACGAGACCTCAGCAAGGACCTGGCGATCGCGTCCTCGGACATCGTGTCCACCTTGCAGGAACGCGGCTTAATGAAATACTGGAAGGGAAAACACATCGTGCTGAAAAAACAGGAGGTGCTAGAGGAGGCGAGCCGGCGCGCCTCCAGAGCTCGCAGCGTTGAGCCTTCCTGTCTGCGGTGGTGGAGCTCGGCCCGCTGA
>DPOGS202818-PA
MISLEWNPYTDVETAGSGSTNPGLPPVIDWEEKNSFLNYNVSCILTLPPYQRQGYGRLLIDFSYLLTKVEGKVGSPETPLSDLGLISYRSYWKEALLKRLCSASGSTLCIRDLSKDLAIASSDIVSTLQERGLMKYWKGKHIVLKKQEVLEEASRRASRARSVEPSCLRWWSSAR-