| DPOGS202991 | ||
|---|---|---|
| Transcript | DPOGS202991-TA | 1080 bp |
| Protein | DPOGS202991-PA | 359 aa |
| Genomic position | DPSCF300068 - 297213-302551 | |
| RNAseq coverage | 3739x (Rank: top 3%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL007279 | 8e-165 | 81.63% | |
| Bombyx | BGIBMGA012336-TA | 1e-156 | 77.04% | |
| Drosophila | ImpL3-PA | 1e-126 | 61.21% | |
| EBI UniRef50 | UniRef50_Q95028 | 2e-124 | 61.21% | L-lactate dehydrogenase n=83 Tax=Coelomata RepID=LDH_DROME |
| NCBI RefSeq | NP_001095933.1 | 3e-155 | 77.04% | lactate dehydrogenase [Bombyx mori] |
| NCBI nr blastp | gi|156255210 | 7e-154 | 77.04% | lactate dehydrogenase [Bombyx mori] |
| NCBI nr blastx | gi|156255210 | 6e-147 | 77.04% | lactate dehydrogenase [Bombyx mori] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0016616 | 8.6e-132 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | |
| GO:0044262 | 8.6e-132 | cellular carbohydrate metabolic process | ||
| GO:0055114 | 8.6e-132 | oxidation-reduction process | ||
| GO:0006096 | 3.9e-116 | glycolysis | ||
| GO:0005737 | 3.9e-116 | cytoplasm | ||
| GO:0004459 | 3.9e-116 | L-lactate dehydrogenase activity | ||
| GO:0003824 | 1.5e-69 | catalytic activity | ||
| GO:0005975 | 1.5e-69 | carbohydrate metabolic process | ||
| GO:0005488 | 8.1e-55 | binding | ||
| GO:0016491 | 6.8e-44 | oxidoreductase activity | ||
| KEGG pathway | cqu:CpipJ_CPIJ014454 | 1e-129 | ||
| K00016 (LDH, ldh) | maps-> | Glycolysis / Gluconeogenesis | ||
| Propanoate metabolism | ||||
| Pyruvate metabolism | ||||
| Cysteine and methionine metabolism | ||||
| InterPro domain | [47-359] IPR001557 | 8.6e-132 | L-lactate/malate dehydrogenase | |
| [52-351] IPR011304 | 3.9e-116 | L-lactate dehydrogenase | ||
| [190-358] IPR015955 | 1.5e-69 | Lactate dehydrogenase/glycoside hydrolase, family 4, C-terminal | ||
| [37-189] IPR016040 | 8.1e-55 | NAD(P)-binding domain | ||
| [48-188] IPR001236 | 6.8e-44 | Lactate/malate dehydrogenase, N-terminal | ||
| [190-352] IPR022383 | 7.6e-31 | Lactate/malate dehydrogenase, C-terminal | ||
| Orthology group | MCL11571 | Multiple-copy universal gene | ||
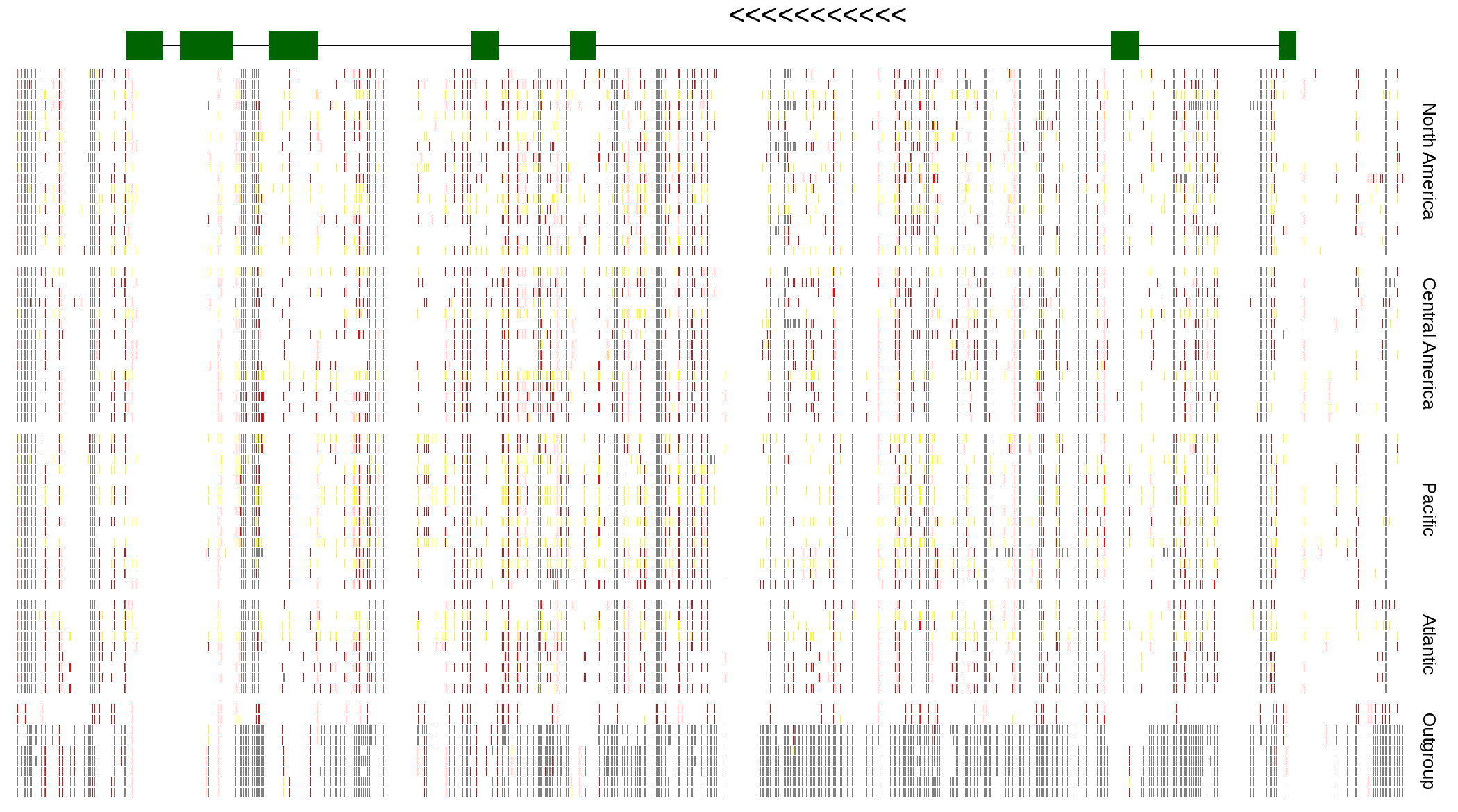
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS202991-TA
ATGAGTTTTATGAACGGGAACGCTAACGGGAATGGGACCGCCAATGGCAACGCTTACAATGGCTTCTACGGTTATAGACACAAAATGGCGACTTTGGACCAGCTTTTCCAGCCCATCCAGCAGAAGACGGAAGCCACCGGCAACAAGGTCACTGTGGTGGGATCTGGCCAAGTCGGCATGGCAGCGGTCTTCGCAATGCTAACACAGGGCGTCACTAACAATATAGCGTTGGTGGATTGTATGGCAGACAAATTGAAAGGCGAAATGATGGATCTACAGCACGGCTCATGCTTCATGAAAGATGCTAAGATCCAGTCAAGCACAGATTACGCCGTCTCTGAGGGTTCTAAGATCTGTGTTGTGACCGCTGGTGTGAGACAACGCGTTGGGGAGACCAGGCTCGAATTGGTACAGAGGAACACTGACGTCTTGAAAGTTATTATACCCCAGCTTGTGAAGTACTGCCCGGACGCCATATTCATCATAGCGAGTAACCCAGTGGACGTTCTAACCTACGTGACCTGGAAAATCAGCGGACTGCCCAAGAACAGGATTATAGGATCCGGAACCAACCTTGACTCAGCTCGTTTCAGGTACTTGCTATCCGAGAAGTTGGCTGTAGCCTCTACTTCCTGTCACGGTTATGTCATCGGTGAACACGGGGACAGCAGTGTACCGGTATGGTCCGGCGTGAACGTAGCCGGTGTGCGTCTCAGCGATCTCAACCCAAAAATAGGCTCGGATGACGATCCTGAAAACTGGAAAAAGATCCACGAAGATGTGGTTAAAAGCGCCTACGAAATCATCAAACTCAAAGGCTATACTTCCTGGGCTATTGGACTGTCTCTATCACAACTCTGTCGCGCTATACTTTACAACATGAATAGCGTGCACCCTGTCACGACCTGCGTTAAGGGCGAGCATGGTATAGAGGACGAAGTGTTCCTGTCCCTGCCCTGCGTGCTCGGCAGGAAAGGCATCTATGACATCATTCGACAAACACTCACCGATAGCGAACTGACCCAGCTTCGTAAATCTGCTGAAGTCATGGCGAAATTGCAAGCCGGTATCAAATTTTAG
>DPOGS202991-PA
MSFMNGNANGNGTANGNAYNGFYGYRHKMATLDQLFQPIQQKTEATGNKVTVVGSGQVGMAAVFAMLTQGVTNNIALVDCMADKLKGEMMDLQHGSCFMKDAKIQSSTDYAVSEGSKICVVTAGVRQRVGETRLELVQRNTDVLKVIIPQLVKYCPDAIFIIASNPVDVLTYVTWKISGLPKNRIIGSGTNLDSARFRYLLSEKLAVASTSCHGYVIGEHGDSSVPVWSGVNVAGVRLSDLNPKIGSDDDPENWKKIHEDVVKSAYEIIKLKGYTSWAIGLSLSQLCRAILYNMNSVHPVTTCVKGEHGIEDEVFLSLPCVLGRKGIYDIIRQTLTDSELTQLRKSAEVMAKLQAGIKF-