| DPOGS204446 | ||
|---|---|---|
| Transcript | DPOGS204446-TA | 477 bp |
| Protein | DPOGS204446-PA | 158 aa |
| Genomic position | DPSCF300002 + 75300-75776 | |
| RNAseq coverage | 11x (Rank: top 84%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL017405 | 4e-71 | 93.67% | |
| Bombyx | BGIBMGA013643-TA | 1e-68 | 87.42% | |
| Drosophila | ato-PA | 1e-25 | 70.37% | |
| EBI UniRef50 | UniRef50_F4WVS5 | 2e-25 | 54.05% | Protein atonal n=2 Tax=Myrmicinae RepID=F4WVS5_ACREC |
| NCBI RefSeq | XP_001121247.1 | 2e-27 | 77.03% | PREDICTED: similar to Protein atonal [Apis mellifera] |
| NCBI nr blastp | gi|379699006 | 7e-66 | 87.42% | atonal [Bombyx mori] |
| NCBI nr blastx | gi|379699006 | 4e-73 | 87.42% | atonal [Bombyx mori] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005634 | 4.2e-22 | nucleus | |
| GO:0006355 | 4.2e-22 | regulation of transcription, DNA-dependent | ||
| KEGG pathway | ||||
| InterPro domain | [102-158] IPR011598 | 4.2e-22 | Helix-loop-helix DNA-binding | |
| [102-153] IPR001092 | 8.4e-17 | Helix-loop-helix DNA-binding domain | ||
| Orthology group | MCL18395 | Insect specific | ||
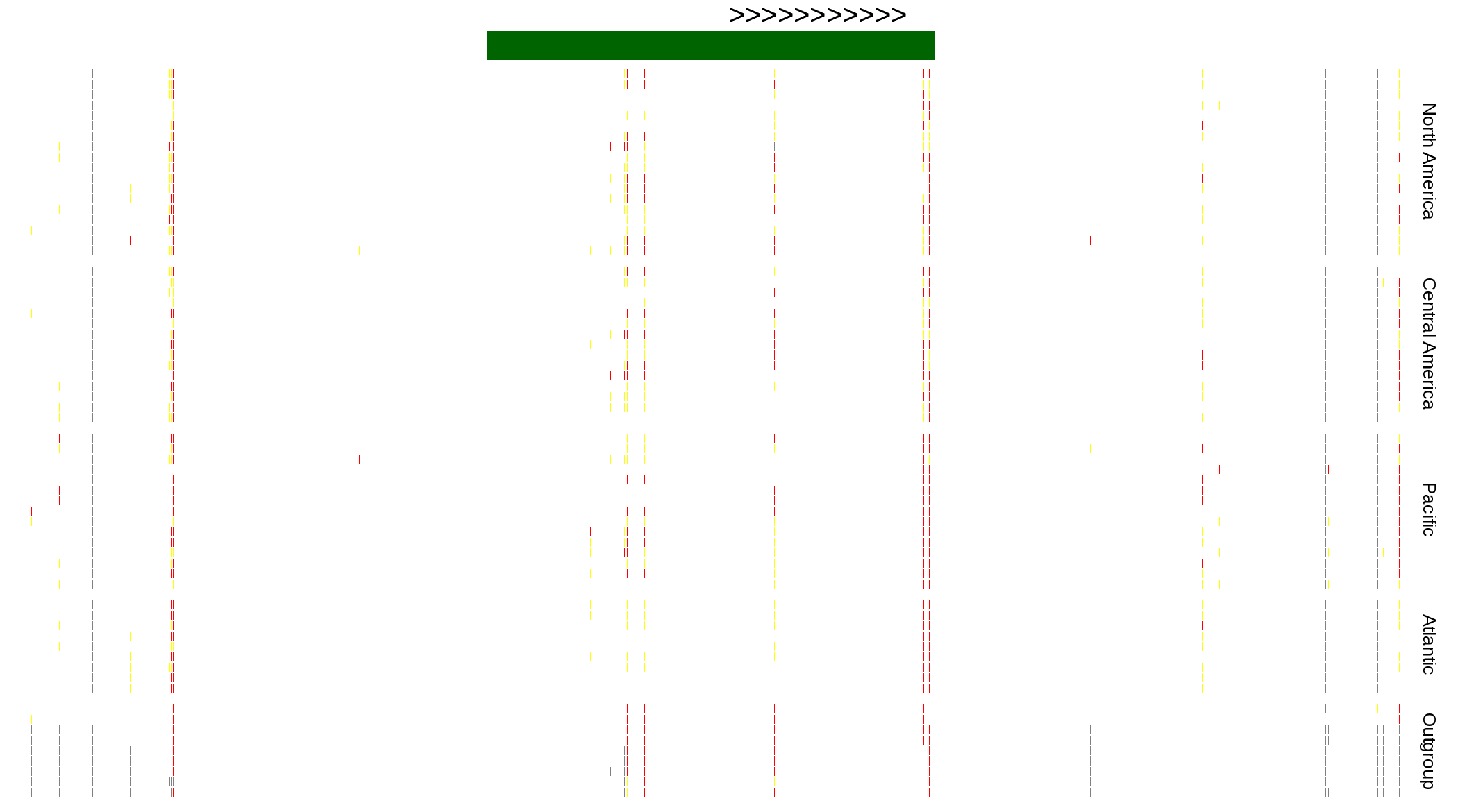
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS204446-TA
ATGGCCGCCGAAACTTACGGACATCGACTGCTCTACACCGAAAAAGATATTTTCCCCAATGACGTTATGTTGGAATATTCTACTGAAGAGTGTTACTTATCCTGGCCAAGGTCACCCGACTCCGGGAGATCCAGCTTGGAGCCAACGCCATCCATGGATGGAAGCAACCAGGAATCACCAACCCACATCGCCTATAGAGGAATGCCGAACGATTTAGTTTTAGAAGACAGTTCGGAAGATTTGGATCTAGAAGGTTCTGGAAAGAGACGAGGAAGGGCGACCAGTGCTGCCGTTTTGCGAAGACGTCGCCTTGCAGCAAACGCGAGGGAAAGAAGACGGATGCAGAACCTGAATAAGGCTTTCGACAGACTACGCGGTCATCTGCCATCGCTCGGAGCCGACAGGCAGCTCTCCAAATACGAGACCCTGCAAATGGCCCAAACATACATCGCCGCATTATACGAGCTGCTACAATAG
>DPOGS204446-PA
MAAETYGHRLLYTEKDIFPNDVMLEYSTEECYLSWPRSPDSGRSSLEPTPSMDGSNQESPTHIAYRGMPNDLVLEDSSEDLDLEGSGKRRGRATSAAVLRRRRLAANARERRRMQNLNKAFDRLRGHLPSLGADRQLSKYETLQMAQTYIAALYELLQ-