| DPOGS206359 | ||
|---|---|---|
| Transcript | DPOGS206359-TA | 4095 bp |
| Protein | DPOGS206359-PA | 1364 aa |
| Genomic position | DPSCF300082 + 1225531-1238842 | |
| RNAseq coverage | 1102x (Rank: top 11%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL017120 | 0.0 | 73.20% | |
| Bombyx | BGIBMGA014119-TA | 0.0 | 77.52% | |
| Drosophila | ade3-PA | 0.0 | 50.66% | |
| EBI UniRef50 | UniRef50_Q26255 | 0.0 | 55.34% | Trifunctional purine biosynthetic protein adenosine-3 n=605 Tax=cellular organisms RepID=PUR2_CHITE |
| NCBI RefSeq | XP_318881.4 | 0.0 | 58.38% | AGAP009786-PA [Anopheles gambiae str. PEST] |
| NCBI nr blastp | gi|158298702 | 0.0 | 58.38% | AGAP009786-PA [Anopheles gambiae str. PEST] |
| NCBI nr blastx | gi|158298702 | 0.0 | 58.42% | AGAP009786-PA [Anopheles gambiae str. PEST] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0009113 | 1.9e-157 | purine base biosynthetic process | |
| GO:0004637 | 1.9e-157 | phosphoribosylamine-glycine ligase activity | ||
| GO:0006189 | 6.7e-126 | 'de novo' IMP biosynthetic process | ||
| GO:0005737 | 6.7e-126 | cytoplasm | ||
| GO:0004641 | 6.7e-126 | phosphoribosylformylglycinamidine cyclo-ligase activity | ||
| GO:0004644 | 7.6e-75 | phosphoribosylglycinamide formyltransferase activity | ||
| GO:0009058 | 5.7e-70 | biosynthetic process | ||
| GO:0016742 | 5.7e-70 | hydroxymethyl-, formyl- and related transferase activity | ||
| GO:0005524 | 2.5e-58 | ATP binding | ||
| GO:0016874 | 2.5e-58 | ligase activity | ||
| GO:0003824 | 1.2e-48 | catalytic activity | ||
| KEGG pathway | aga:AgaP_AGAP009786 | 0.0 | ||
| K11787 (GART) | maps-> | Purine metabolism | ||
| One carbon pool by folate | ||||
| InterPro domain | [4-428] IPR000115 | 1.9e-157 | Phosphoribosylglycinamide synthetase | |
| [801-1134] IPR004733 | 6.7e-126 | Phosphoribosylformylglycinamidine cyclo-ligase | ||
| [106-299] IPR020561 | 2.8e-86 | Phosphoribosylglycinamide synthetase, ATP-grasp (A) domain | ||
| [1163-1355] IPR004607 | 7.6e-75 | Phosphoribosylglycinamide formyltransferase | ||
| [1163-1364] IPR002376 | 5.7e-70 | Formyl transferase, N-terminal | ||
| [195-331] IPR013816 | 2.5e-58 | ATP-grasp fold, subdomain 2 | ||
| [969-1144] IPR010918 | 2.3e-50 | AIR synthase-related protein, C-terminal | ||
| [799-967] IPR016188 | 1.2e-48 | PurM, N-terminal-like | ||
| [4-105] IPR020562 | 4.1e-36 | Phosphoribosylglycinamide synthetase, N-terminal | ||
| [1-106] IPR016185 | 1.8e-33 | PreATP-grasp-like fold | ||
| [4-98] IPR013817 | 1.1e-32 | Pre-ATP-grasp fold | ||
| [335-428] IPR020560 | 2.6e-26 | Phosphoribosylglycinamide synthetase, C-domain | ||
| [332-430] IPR011054 | 2.5e-25 | Rudiment single hybrid motif | ||
| [124-194] IPR013815 | 3.2e-20 | ATP-grasp fold, subdomain 1 | ||
| [481-572] IPR000728 | 1.6e-17 | AIR synthase-related protein | ||
| Orthology group | MCL14235 | Single-copy universal gene | ||
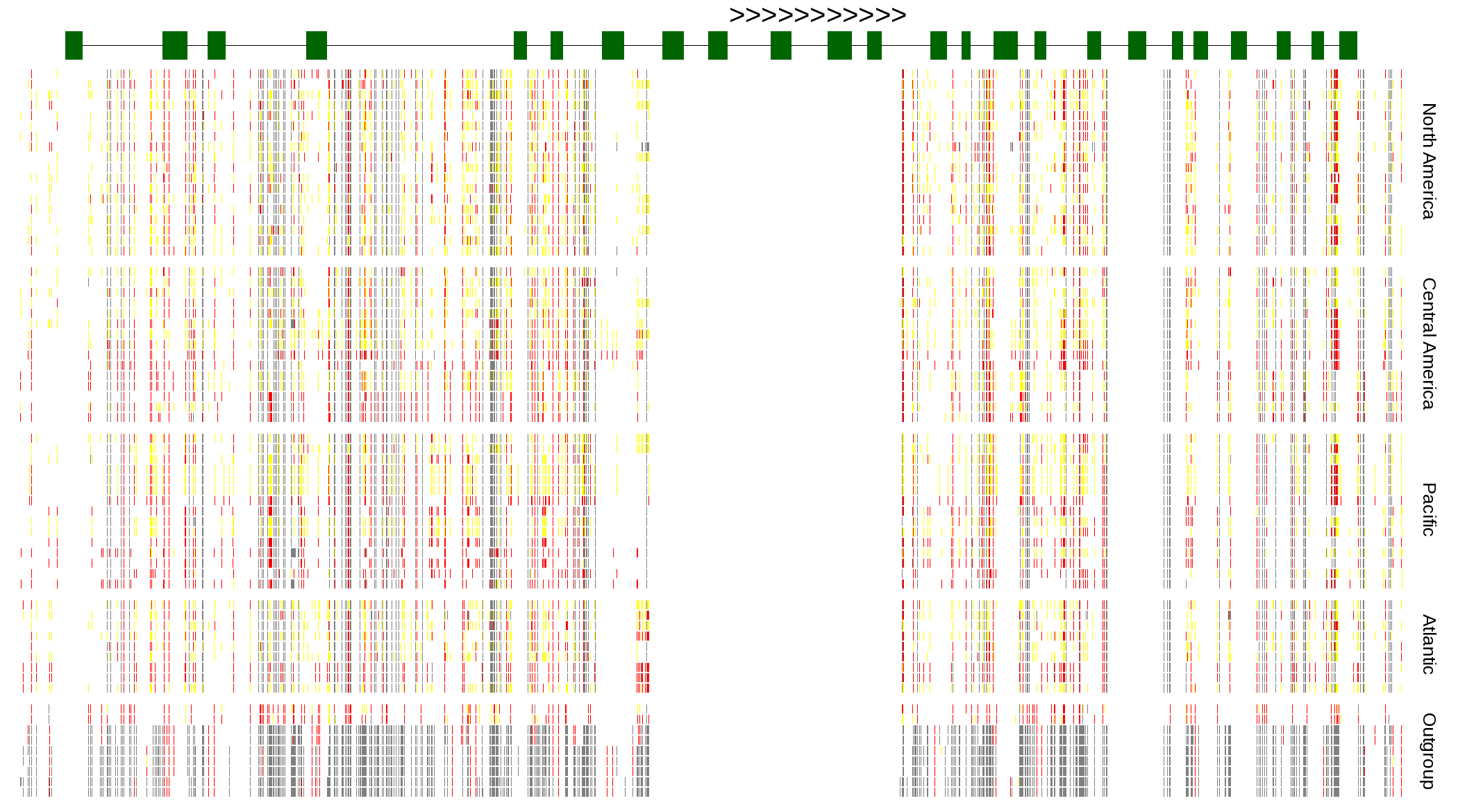
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS206359-TA
ATGTCGGCAAACGTGCTCGTTATCGGTGGTGGAGGCAGGGAACATGCAATTTGTTGGAAATTAGCAGATTCTCCATTAATTTGTAAAATATTCTGTGCGCCCGGTAGTGTCGGTATATCGGCGACGAAGGAAAATGTTGAATGTGTGGATTTGAATATTAAGGATTTTCCGGGTCTGGCGAAGTGGTGCAAAGACAAGTCAATTGATCTCGTCATCATTGGACCCGAAGACCCTCTTGCTAATGGCATTGTGGATGCCCTCGAACCAGCTGGCATCAAATGCTTCGGACCCACAAAAGCCGGTGCACAGATCGAGGCCAATAAAGACTGGTCCAAGAAATTTATGAACAAATATCAAATACCAACGGCAAGATACCAATCATTCACAGATGCTGAAGCTGCTAAGCAATTCATTAAGAGTGCACCGTTTAGAGCGTTAGTCGTGAAAGCATCCGGTTTAGCGGCTGGGAAAGGAGTTGTGGTAGCGAGCAATGTAGATGAGGCCTGTGCAGCGGTGGATGAGATCTTAACCGAAGCCAAGTACGGAACTGCCGGGCAGGTGGTCGTGGTTGAAGAACTGTTGGAAGGGGAAGAGGTTTCGGTGCTGGCATTCACGGATGGGAACACGGTGTCCATGATGCCCCCCGCCCAGGATCACAAGCGCATCGGCGAGGGGGACACGGGACCCAACACGGGGGGTATGGGGGCATATTGTCCCTGTCCGCTTATCACCCCAGAACAGCTGGCCGATGTCAAGGATCAAGTGTTACAGAGAGCTGTGGACGGGCTCAGGGCTGAGGGCATCAAATATGTCGGGGTCCTCTACGCTGGTCTTATGGTGACCAAGTCTGGTCCAATGACCCTCGAGTTTAACTGCCGCTTCGGAGACCCAGAGACACAGGTCCTCATGATGTTGCTGGAATCGGATCTCTACTCCGTCATTAAGGCGTGTGTGAGCGGTACATTAAAGGAGACTCCGGTCAAATGGAACACCTCAATGTCGGCCGTGGGAGTGGTGATCGCTTCCAAAGGTTACCCTGAGAGCTCCACCAAGGGCTGTGTCATAAGCGGTCTATCCCAGGTGTCCAGGGAAGACGTGGTCGTGTTCATGAGCGGAGTGTCCCGTGGAGCAAACGACTCGCTGGTCACTGCCGGCGGCAGAGTGCTGCTGCTGGCGGCGAGGAGAGCCGACCTCCGGACTGCCGCCGCCGCTGCGACACGAGCCGCCGCCGCCGTGGACTTCCCCGGGAAACAGTACAGGAAGGACATCGCCAGGCGGGCCTTCTGCAAAATGAACGGCCTGTCATACCTGGAGAGCGGTGTGGACATTGAAGCGGCGGCGGCCTTGGTCCGTCTGATGGAGCCCCTGGCCACGGGGACCCACAGACCCGGGGTACTTGGAAGGCTGGGCTGCTACAGCGGACTGTTCCAACTGGCGGCCGTGGACCCCGGCCTCACAGACCCCGTGCTGGTCCAGGGAACGGACGGCGTTGGAACCAAGGTCAAGATAGCAGAGATGATGCAGAAGTACGACACCATCGGCCAGGACCTGGTGGCCATGTGCGTCAACGACATCCTGTGTGCGGGCGCCGAGCCCTTCGCCTTCCTGGACTACCTGGCGTGCGGCCGGCTGCAGCTGCACACCTCCACCACCATCGTCAAGGGAATCGCCGACGCCTGCGTCATGGCCGGCTGTGCTTTGTTGGGGGGAGAGACGGCGGAGATGCCGAGTATGTATGACGTGGGGAAGTACGATTTGGCGGGGTTCGCGGTGGGCGTGGTGGACAACCTCAAGCAGCTGCCCCGCTACAAGGAGATACGACCCGGGGACGTGGTGCTCGCGCTGCCCTCCACCGGCGTGCACAGCAACGGGTACAGCCTCGTGCAGAGGATCATGGCTGAAAGTGGACATAGTTTCTACGAAAAGGCTCCGTTCAGTAAATCCAACAAGAACTTCGGCGAGGAGTTCCTGGAGCCGACCGGTATCTACGTGAAGGCCCTCCTGCCGGCCATCAAGAAAGGCCTCGTGAAGGGCCTGGCACACATCACCGGGGGAGGCCTCCTGGAGAATATCCCCAGGATACTACCGCCCGGCGTCAGGGTCAGGCTCGACGCCACTAAGTTCCAGATAAACCCTATCTTCGGCTGGCTGCAAGCTAAAGGGATGGTGTCGGACTTCGAGATGCTGCGCACGTTCAACTGCGGTGTCGGCATGGTGGTGGTGGCGGACCCCGTGCTGGTGAGCGAGCTGGTCGCCGCCGTGGACGGCACCATCAGTGTGGTCGGGCAGCTGGAGGACATGAGAACGGAAGGAGGTCAGCAGGTCATAGTGGAGAACTTCCAGCAGGCCATGTCCCCTCTGACGTCACCCTACTCGTCCGCCAGTCCGTGTCAGAAGTCACTCTCCTACAAGGACAGCGGGGTCGACATCGAGGCCGGGGACTCGCTGGTGTCACTCATAAAGCCTTTGGCTAGATCCACGTCTCGATCCGGGGTCCTGGGAGGTCTTGGAGGCTTCGGCGGGTGTTTCCAGCTGAAGGCTGTGGAGCAGGAGTATAAGGACCCGGTGCTGGTGGTGGCGGCGGACGGTGTGGGCACCAAGCTGCGCGTGGCTCAGAAGATGAACCGACACGCCACCATCGGCGTTGACCTGGTGGCCATGTGCGTCAACGACATCCTGTGCAACGGCGCCGCGCCGCTCACCTTCCTGGACTACTTCGCCTGCGGAGCCCTGGACGTGACCGTGGCCAGGGACGTCGTGGCCGGGGTCGCGGACGGCTGCAAGCAGTCCTCAGCGGCTCTCATCGGCGGAGAGACGGCGGAGATGCCCGGCATGTACGAGGCCGGCGTGTACGACATCGCAGGGTTCGCGCTGGGAGTGGTGGAGAGGGACAACATACTGCCGAAGATCAACGACATCAATGTTGGCGACACGATAATAGGTCTGCCATCGAACGGCGTCCACAGCAACGGGTTCAGTCTCATCCACAGCCTCATGAAGAAGGCCGGTCTGAGTCTCAACGACAAGGCGCCCTTCAGCGAGGAAGGACTCACTCTCGGCCAGGAGCTGATCAAGCCGACCCGCATCTACGTCCGCAGCGTGCTGCCGGCGCTGCAGCGCGGCGTGGTGAAGGCGGTGGCGCACATCACGGGCGGCGGCCTCATGGAGAACATCCCGCGCATCATGCCGGACTCCGTGCGGGCCCGCCTCAACGCGCACTGGTGGAAAGTTCACCCTGTGTTCGCGTGGATCGCGGAGACCGGCGAGGTCAAGAACGACGAGATGCTGAGGACATTCAACTGCGGCATCGGCTTGGTGCTGATAGTGTCTCCGGAACACCAGGCAGAGGTGATGAACATCACTCGCTCGCACGGCGCGATGGTGATCGGCTCCATACAAGCCCGGCCCCCGGGCGGCGCTCGCGTGCTCGTCGACAACTTCACCTCCGCGCTGGACTTCACGAGGCGGATGCCGCACCTCACTAAGAAGAGGGTGGCGGTGCTGGTGTCGGGTAGCGGCAGCAACCTGCAGGCGCTCATGGACAGTGCGTCGGACCCCGCCCAGTGCATGTGTGCGGAGGTGGCGCTCGTCGTCAGCAACAAACCCGACGCCTTCGCCCTCAAACGGGCCCGGGACGCCGGCGTCAACACGCTGGTGCTGAGTCACAAGGACTACTCCAGCCGCGAGGAGTACGACCGCGCCCTCAGCGCCGCCCTGGACGCGCACCGGATCGACCTCGTGTGTCTGGCCGGCTTCATGAGGATACTCACGCCGGGCTTCGTTAAGAAGTGGAAGGGTCGCCTCATCAACATCCACCCGTCCCTGCTGCCGGCCCACCCTGGACTCCACGCTCAGAGACAGTGTCTACAGGCGGGAGACAAGGAGTCGGGCTGCACCGTACACTTCGTCGACGAGGGCATGGACACGGGTCCGATCATTCTCCAGGAGCGCGTGCCGGTGATGCCGGGAGACACGGAGCAGGTTCTCAGTGACAGGATCCTGTCCGCGGAACACCGCGCCTACCCTCAGGCGCTCAGACTGCTCGCTACGGGCCGGGTCCGGCTACATGAGGACACTATCATATGGCATTCATGA
>DPOGS206359-PA
MSANVLVIGGGGREHAICWKLADSPLICKIFCAPGSVGISATKENVECVDLNIKDFPGLAKWCKDKSIDLVIIGPEDPLANGIVDALEPAGIKCFGPTKAGAQIEANKDWSKKFMNKYQIPTARYQSFTDAEAAKQFIKSAPFRALVVKASGLAAGKGVVVASNVDEACAAVDEILTEAKYGTAGQVVVVEELLEGEEVSVLAFTDGNTVSMMPPAQDHKRIGEGDTGPNTGGMGAYCPCPLITPEQLADVKDQVLQRAVDGLRAEGIKYVGVLYAGLMVTKSGPMTLEFNCRFGDPETQVLMMLLESDLYSVIKACVSGTLKETPVKWNTSMSAVGVVIASKGYPESSTKGCVISGLSQVSREDVVVFMSGVSRGANDSLVTAGGRVLLLAARRADLRTAAAAATRAAAAVDFPGKQYRKDIARRAFCKMNGLSYLESGVDIEAAAALVRLMEPLATGTHRPGVLGRLGCYSGLFQLAAVDPGLTDPVLVQGTDGVGTKVKIAEMMQKYDTIGQDLVAMCVNDILCAGAEPFAFLDYLACGRLQLHTSTTIVKGIADACVMAGCALLGGETAEMPSMYDVGKYDLAGFAVGVVDNLKQLPRYKEIRPGDVVLALPSTGVHSNGYSLVQRIMAESGHSFYEKAPFSKSNKNFGEEFLEPTGIYVKALLPAIKKGLVKGLAHITGGGLLENIPRILPPGVRVRLDATKFQINPIFGWLQAKGMVSDFEMLRTFNCGVGMVVVADPVLVSELVAAVDGTISVVGQLEDMRTEGGQQVIVENFQQAMSPLTSPYSSASPCQKSLSYKDSGVDIEAGDSLVSLIKPLARSTSRSGVLGGLGGFGGCFQLKAVEQEYKDPVLVVAADGVGTKLRVAQKMNRHATIGVDLVAMCVNDILCNGAAPLTFLDYFACGALDVTVARDVVAGVADGCKQSSAALIGGETAEMPGMYEAGVYDIAGFALGVVERDNILPKINDINVGDTIIGLPSNGVHSNGFSLIHSLMKKAGLSLNDKAPFSEEGLTLGQELIKPTRIYVRSVLPALQRGVVKAVAHITGGGLMENIPRIMPDSVRARLNAHWWKVHPVFAWIAETGEVKNDEMLRTFNCGIGLVLIVSPEHQAEVMNITRSHGAMVIGSIQARPPGGARVLVDNFTSALDFTRRMPHLTKKRVAVLVSGSGSNLQALMDSASDPAQCMCAEVALVVSNKPDAFALKRARDAGVNTLVLSHKDYSSREEYDRALSAALDAHRIDLVCLAGFMRILTPGFVKKWKGRLINIHPSLLPAHPGLHAQRQCLQAGDKESGCTVHFVDEGMDTGPIILQERVPVMPGDTEQVLSDRILSAEHRAYPQALRLLATGRVRLHEDTIIWHS-