| DPOGS207161 | ||
|---|---|---|
| Transcript | DPOGS207161-TA | 336 bp |
| Protein | DPOGS207161-PA | 111 aa |
| Genomic position | DPSCF300001 + 4517689-4518314 | |
| RNAseq coverage | 4x (Rank: top 89%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL013045 | 2e-17 | 53.57% | |
| Bombyx | % | |||
| Drosophila | % | |||
| EBI UniRef50 | % | |||
| NCBI RefSeq | XP_001599730.1 | 1e-07 | 39.08% | PREDICTED: similar to activin receptor type I, putative [Nasonia vitripennis] |
| NCBI nr blastp | gi|332021727 | 2e-07 | 41.67% | TGF-beta receptor type-1 [Acromyrmex echinatior] |
| NCBI nr blastx | gi|332021727 | 3e-07 | 41.67% | TGF-beta receptor type-1 [Acromyrmex echinatior] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0016020 | 7.5e-10 | membrane | |
| GO:0005024 | 7.5e-10 | transforming growth factor beta receptor activity | ||
| GO:0004675 | 7.5e-10 | transmembrane receptor protein serine/threonine kinase activity | ||
| KEGG pathway | dre:30183 | 1e-07 | ||
| K13567 (ACVR1B) | maps-> | Cytokine-cytokine receptor interaction | ||
| InterPro domain | [6-83] IPR000472 | 7.5e-10 | TGF-beta receptor/activin receptor, type I/II | |
| Orthology group | MCL26647 | Insect specific | ||
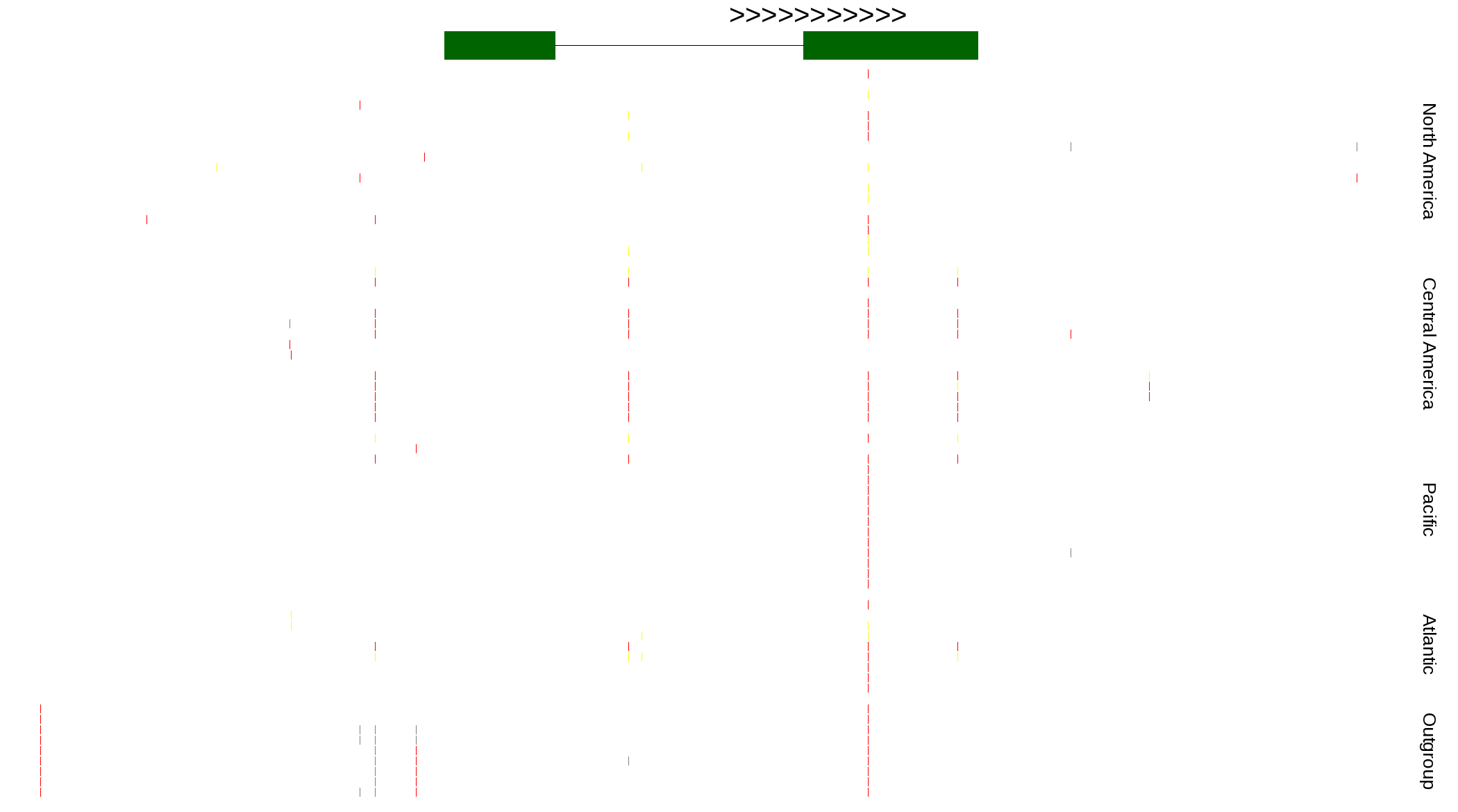
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS207161-TA
ATGGTTGTGGCAGGATTGAAATGTTACTGTAACAGTGACAGTCCCAGCTGCCCAAACTCCACCTGTGAGACAGATGGCTTCTGTATGGCATCTACCAGCATTGAGAATGGTGAACATAAGCGCACCTACCAAGTGGTGGCCTTAAGCTGTTTGCAAAGCAATGCATTCCCACCAGAGAATCCAATCAAATGCAAAAACCCGAGAACGTTTTGCTGTAAAAATGATTTCTGCAACTCTGATGAGGTTTACCAAGAGAGATTAACAAAAGTGCTCAATGCCTCAAGCTCAGCCTCTGGTAAGGGATGCTCACGCACTTGTGGCGTGTATACAAACTGA
>DPOGS207161-PA
MVVAGLKCYCNSDSPSCPNSTCETDGFCMASTSIENGEHKRTYQVVALSCLQSNAFPPENPIKCKNPRTFCCKNDFCNSDEVYQERLTKVLNASSSASGKGCSRTCGVYTN-