| DPOGS207468 | ||
|---|---|---|
| Transcript | DPOGS207468-TA | 675 bp |
| Protein | DPOGS207468-PA | 224 aa |
| Genomic position | DPSCF300051 + 59012-59939 | |
| RNAseq coverage | 121x (Rank: top 57%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL016684 | 8e-118 | 87.05% | |
| Bombyx | BGIBMGA001174-TA | 1e-112 | 84.82% | |
| Drosophila | CG32485-PA | 4e-87 | 67.26% | |
| EBI UniRef50 | UniRef50_Q8I941 | 5e-85 | 67.26% | CG32485 n=14 Tax=Diptera RepID=Q8I941_DROME |
| NCBI RefSeq | XP_002426588.1 | 3e-94 | 74.65% | SEC14 cytosolic factor, putative [Pediculus humanus corporis] |
| NCBI nr blastp | gi|332374646 | 3e-94 | 73.18% | unknown [Dendroctonus ponderosae] |
| NCBI nr blastx | gi|332374646 | 4e-93 | 73.18% | unknown [Dendroctonus ponderosae] |
| Group | ||||
|---|---|---|---|---|
| KEGG pathway | uma:UM04979.1 | 1e-08 | ||
| K01101 (E3.1.3.41) | maps-> | gamma-Hexachlorocyclohexane degradation | ||
| InterPro domain | [61-223] IPR001251 | 4.2e-41 | Cellular retinaldehyde-binding/triple function, C-terminal | |
| [3-73] IPR011074 | 5.8e-10 | Phosphatidylinositol transfer protein-like, N-terminal | ||
| Orthology group | MCL18946 | Insect specific | ||
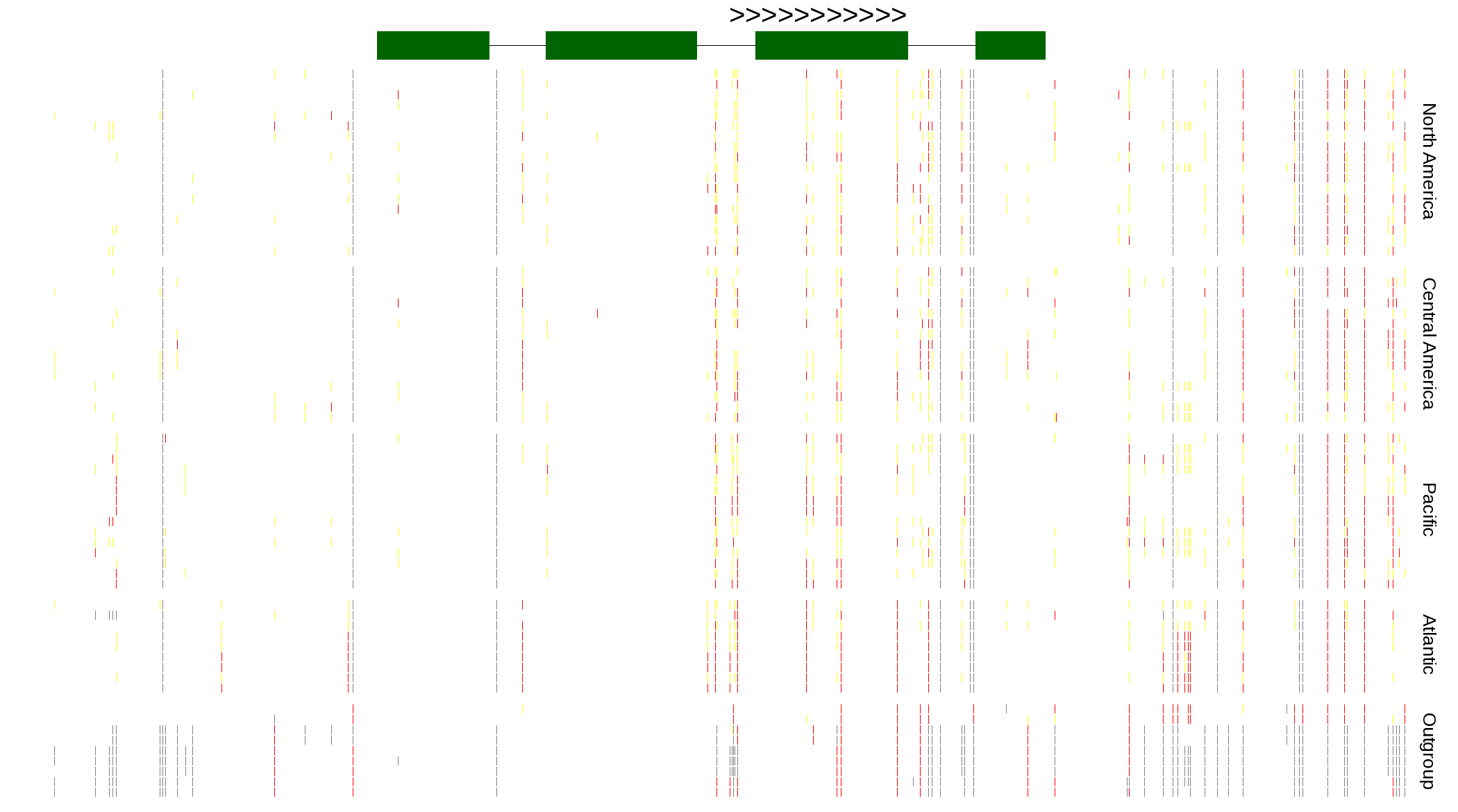
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS207468-TA
ATGGCAGAAGAATTAAAACCTGTTAAAGACGAAGACCTAAAACAACTCAAAGAACGTATGCAACTAATTGCTGAGGCAGATCCATCACAATTCCATAATGATTATTCCTTGAAGAGATACTTAAGGGCTTTCAAAACTGTGGATAATGCATTTCAGGCTATTTTAAAAAGTAACAAATGGAGAGTTGAATACGGTGTAGCCAATTTGCATGAAAACCATGAATTAATTGAGAAATATTCTAATCGAGCTAGAGTTCTTCGTCACAGAGATATGATTGGGAGACCAATTGTATACATTCCAGCCAAAAACCATAGCTCCAGTGATAGAAGTATTGATGAGCTTACAAAGTTTATTGTCTACTGCTTAGAAGACGCAAGTAAGAAATGTTTTGAAGAAGTTATTGACAATCTTTGTATTGTTTTTGACCTTAATAATTTCACATTATCTTGCATGGACTACCAAGTTTTAAAAAATCTCATTTGGCTTCTTAGCCGGCATTATCCAGAAAGACTTGGCGTTTGCCTCATTATAAATGCACCAACATTTTTCTCTGGCTGCTGGGCTGTTATAAAAGGATGGCTGGATGAAAATACTGCTGGCAAAGTTACTTTTGTCAACTCGGAAATGGATCTTTGTCAGTACCTGATACCAGATATACTACCAACTGATATGTAA
>DPOGS207468-PA
MAEELKPVKDEDLKQLKERMQLIAEADPSQFHNDYSLKRYLRAFKTVDNAFQAILKSNKWRVEYGVANLHENHELIEKYSNRARVLRHRDMIGRPIVYIPAKNHSSSDRSIDELTKFIVYCLEDASKKCFEEVIDNLCIVFDLNNFTLSCMDYQVLKNLIWLLSRHYPERLGVCLIINAPTFFSGCWAVIKGWLDENTAGKVTFVNSEMDLCQYLIPDILPTDM-