| DPOGS208422 | ||
|---|---|---|
| Transcript | DPOGS208422-TA | 204 bp |
| Protein | DPOGS208422-PA | 67 aa |
| Genomic position | DPSCF300390 - 51113-51739 | |
| RNAseq coverage | 172x (Rank: top 50%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL016262 | 2e-16 | 78.72% | |
| Bombyx | BGIBMGA007441-TA | 3e-18 | 64.91% | |
| Drosophila | Inos-PA | 3e-16 | 74.47% | |
| EBI UniRef50 | UniRef50_O97477 | 6e-14 | 74.47% | Inositol-3-phosphate synthase n=10 Tax=Eukaryota RepID=INO1_DROME |
| NCBI RefSeq | XP_001986082.1 | 7e-15 | 74.47% | GH20732 [Drosophila grimshawi] |
| NCBI nr blastp | gi|332374410 | 7e-14 | 78.72% | unknown [Dendroctonus ponderosae] |
| NCBI nr blastx | gi|332374410 | 9e-13 | 78.72% | unknown [Dendroctonus ponderosae] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008654 | 4.2e-21 | phospholipid biosynthetic process | |
| GO:0006021 | 4.2e-21 | inositol biosynthetic process | ||
| GO:0004512 | 4.2e-21 | inositol-3-phosphate synthase activity | ||
| GO:0005488 | 2.7e-15 | binding | ||
| KEGG pathway | tca:655540 | 2e-14 | ||
| K01858 (E5.5.1.4, INO1) | maps-> | Inositol phosphate metabolism | ||
| Streptomycin biosynthesis | ||||
| InterPro domain | [22-60] IPR002587 | 4.2e-21 | Myo-inositol-1-phosphate synthase | |
| [16-63] IPR016040 | 2.7e-15 | NAD(P)-binding domain | ||
| Orthology group | ||||
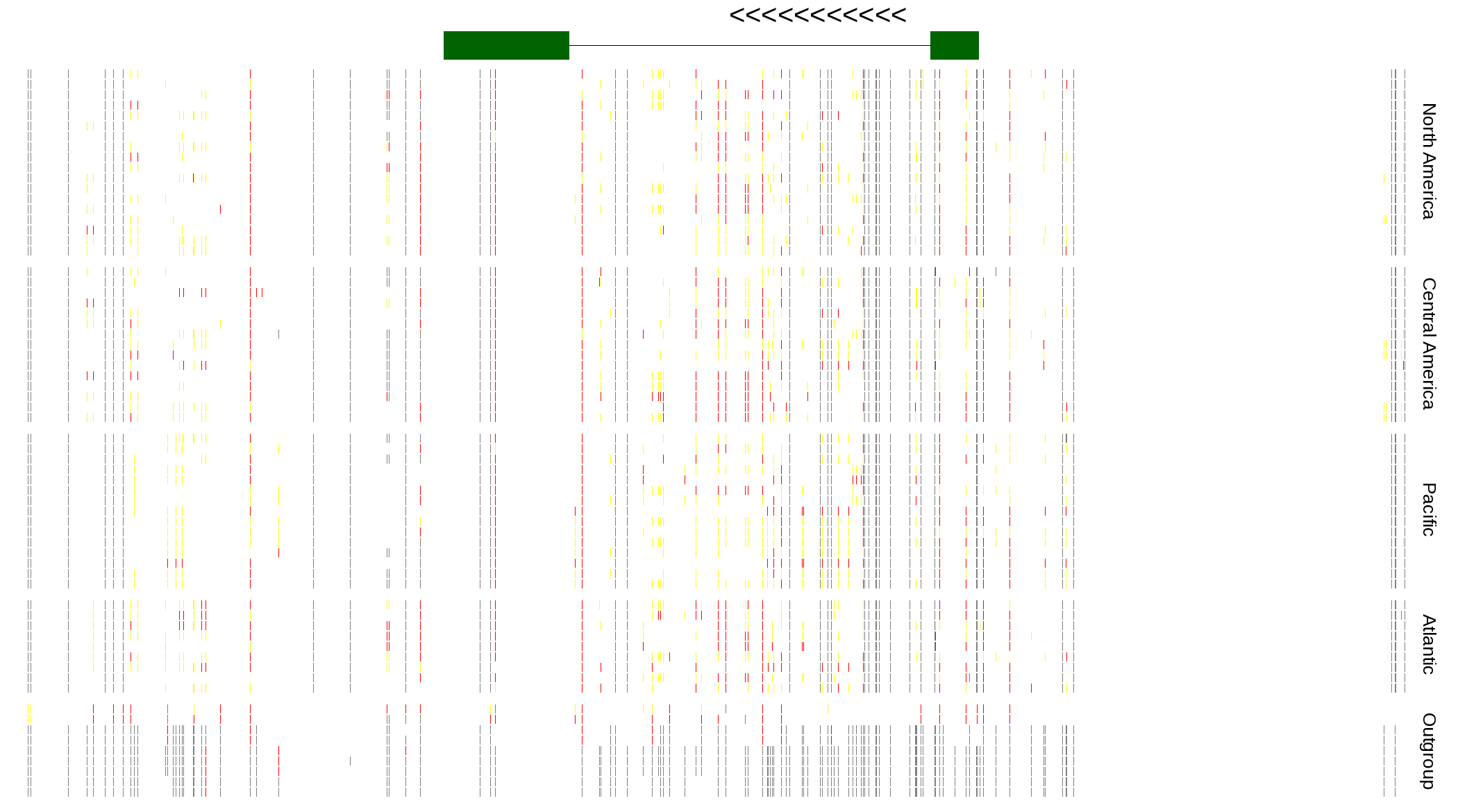
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS208422-TA
ATGATTTTCTCTGCTTACAGAGGAATTGTTGTTGTCTGTGAGAGTGCAGAAGTGAAGTGTTGCTACATCAACGGCTCTCCTCAGAACACGCTGGTCCCGGGGCTGGTGGAGCTCGCTGAGCGGAAAGGAGTGTTTGTGGCCGGAGACGATTTCAAATCCGGTCAAACCAAGCTCAAATCCCGCTCTGTTAGCGTATTAAGCTAA
>DPOGS208422-PA
MIFSAYRGIVVVCESAEVKCCYINGSPQNTLVPGLVELAERKGVFVAGDDFKSGQTKLKSRSVSVLS-