| DPOGS208471 | ||
|---|---|---|
| Transcript | DPOGS208471-TA | 579 bp |
| Protein | DPOGS208471-PA | 192 aa |
| Genomic position | DPSCF300064 - 1533385-1535930 | |
| RNAseq coverage | 305x (Rank: top 37%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL004298 | 8e-50 | 52.08% | |
| Bombyx | BGIBMGA010653-TA | 2e-69 | 78.23% | |
| Drosophila | mRpL48-PA | 4e-38 | 45.96% | |
| EBI UniRef50 | UniRef50_C4WU99 | 8e-40 | 50.00% | Putative uncharacterized protein n=2 Tax=Acyrthosiphon pisum RepID=C4WU99_ACYPI |
| NCBI RefSeq | XP_001659904.1 | 2e-41 | 49.03% | mitochondrial ribosomal protein, L48, putative [Aedes aegypti] |
| NCBI nr blastp | gi|157121535 | 3e-40 | 49.03% | mitochondrial ribosomal protein, L48, putative [Aedes aegypti] |
| NCBI nr blastx | gi|157121535 | 2e-39 | 49.34% | mitochondrial ribosomal protein, L48, putative [Aedes aegypti] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005840 | 1.5e-10 | ribosome | |
| GO:0006412 | 1.5e-10 | translation | ||
| GO:0005622 | 1.5e-10 | intracellular | ||
| GO:0003735 | 1.5e-10 | structural constituent of ribosome | ||
| KEGG pathway | ||||
| InterPro domain | [57-154] IPR001848 | 1.5e-10 | Ribosomal protein S10 | |
| Orthology group | MCL12910 | Single-copy universal gene | ||
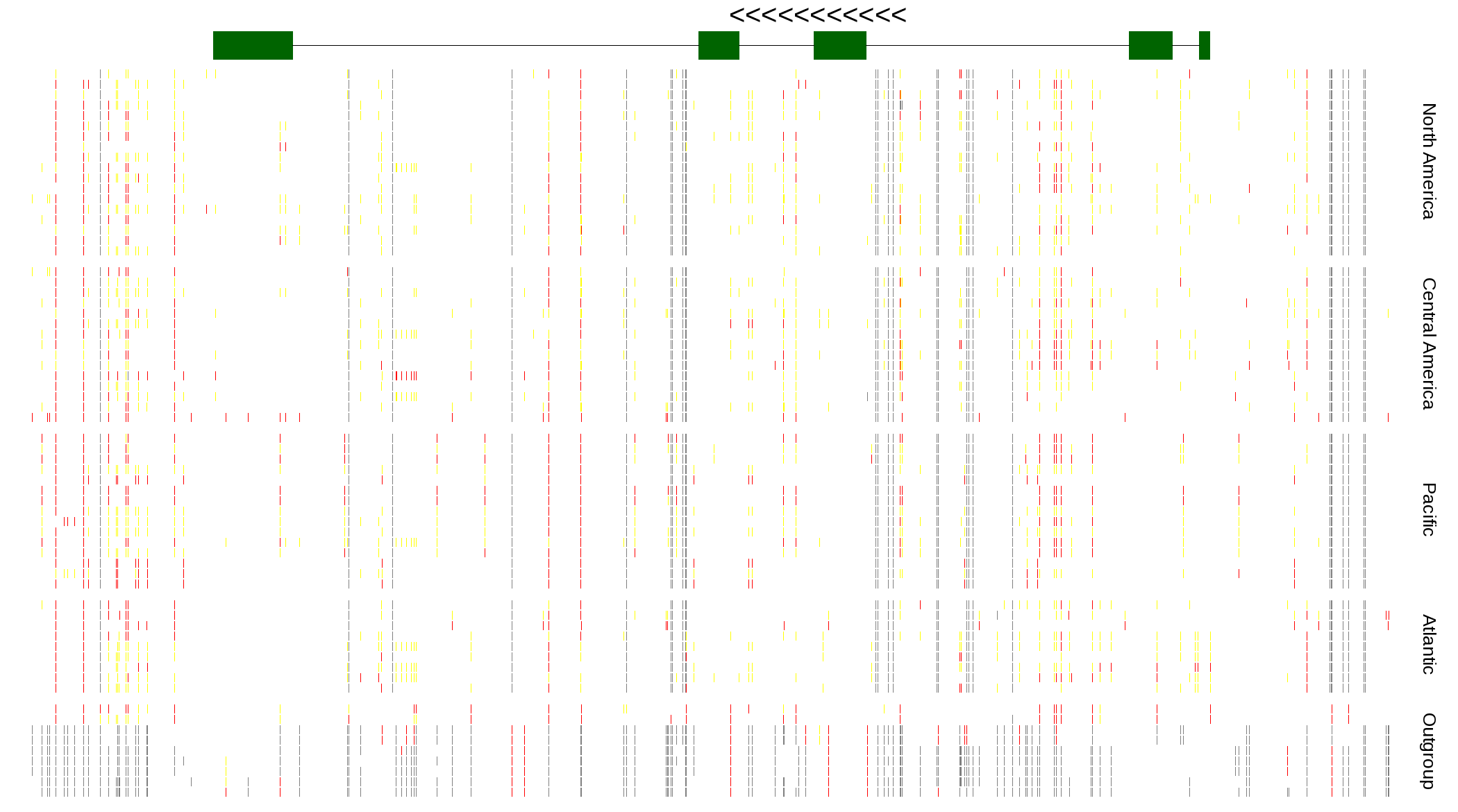
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS208471-TA
ATGTTTAGTTCGAAACTATTAAATATAGTGAAACTAAATCGTTATATCAATCCAAAAACATTTGCCAAACAGAATGTATCATTTCAATGTGCTCAGAAGCGACACAAAAGCGACTTATACGAGCCTGATTACCTCATTAGTATGCAACCCGATGAACCTGTCTATAATTTAATAAACTTGCAACTTAAAAGTTATGACTTCACTCTACTTGAATCATGTCAAAAAGAAATACACAGATATGCTGAAGTAATGGGAATCGAAGTTGAAGATGGTTGGGCAACACCTGCTCAACAATTGAAGATACAAAGATTTAAGCCCAATAGCACAGCTATGGAATCTGAATATCATCTTAATGTTTATGAAAGGAATGTACAGGTGTCGGACGTCCCGGCCTGGGCGCTGGGAACACTTCTTCGAGTTTCGCGCGCTCTTCTTCCCGAAGGTTGCACCCTCAACGTTCACGAACACACACAAGCTCACGATGAAATCAGATACGTACCTGACAACGATTTGATAGAACTCAAACAACAGCTGGAGGAAATGGGCGGCAGTCGCGCGGAACGAAAAAAGAAGAAATAG
>DPOGS208471-PA
MFSSKLLNIVKLNRYINPKTFAKQNVSFQCAQKRHKSDLYEPDYLISMQPDEPVYNLINLQLKSYDFTLLESCQKEIHRYAEVMGIEVEDGWATPAQQLKIQRFKPNSTAMESEYHLNVYERNVQVSDVPAWALGTLLRVSRALLPEGCTLNVHEHTQAHDEIRYVPDNDLIELKQQLEEMGGSRAERKKKK-