| DPOGS208935 | ||
|---|---|---|
| Transcript | DPOGS208935-TA | 585 bp |
| Protein | DPOGS208935-PA | 194 aa |
| Genomic position | DPSCF300009 + 114656-115470 | |
| RNAseq coverage | 651x (Rank: top 20%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL004763 | 1e-106 | 92.75% | |
| Bombyx | BGIBMGA002406-TA | 6e-108 | 95.31% | |
| Drosophila | Jafrac2-PB | 1e-90 | 78.95% | |
| EBI UniRef50 | UniRef50_O08807 | 7e-92 | 81.15% | Peroxiredoxin-4 n=7 Tax=Eutheria RepID=PRDX4_MOUSE |
| NCBI RefSeq | XP_001648972.1 | 3e-96 | 84.21% | peroxiredoxins, prx-1, prx-2, prx-3 [Aedes aegypti] |
| NCBI nr blastp | gi|157674503 | 4e-98 | 88.42% | putative peroxiredoxin [Lutzomyia longipalpis] |
| NCBI nr blastx | gi|157674503 | 3e-97 | 88.42% | putative peroxiredoxin [Lutzomyia longipalpis] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0016209 | 2.2e-36 | antioxidant activity | |
| GO:0055114 | 2.2e-36 | oxidation-reduction process | ||
| GO:0016491 | 2.2e-36 | oxidoreductase activity | ||
| GO:0051920 | 2.4e-15 | peroxiredoxin activity | ||
| KEGG pathway | ||||
| InterPro domain | [1-191] IPR012336 | 4.3e-72 | Thioredoxin-like fold | |
| [2-134] IPR000866 | 2.2e-36 | Alkyl hydroperoxide reductase subunit C/ Thiol specific antioxidant | ||
| [155-190] IPR019479 | 2.4e-15 | Peroxiredoxin, C-terminal | ||
| Orthology group | MCL16648 | Patchy | ||
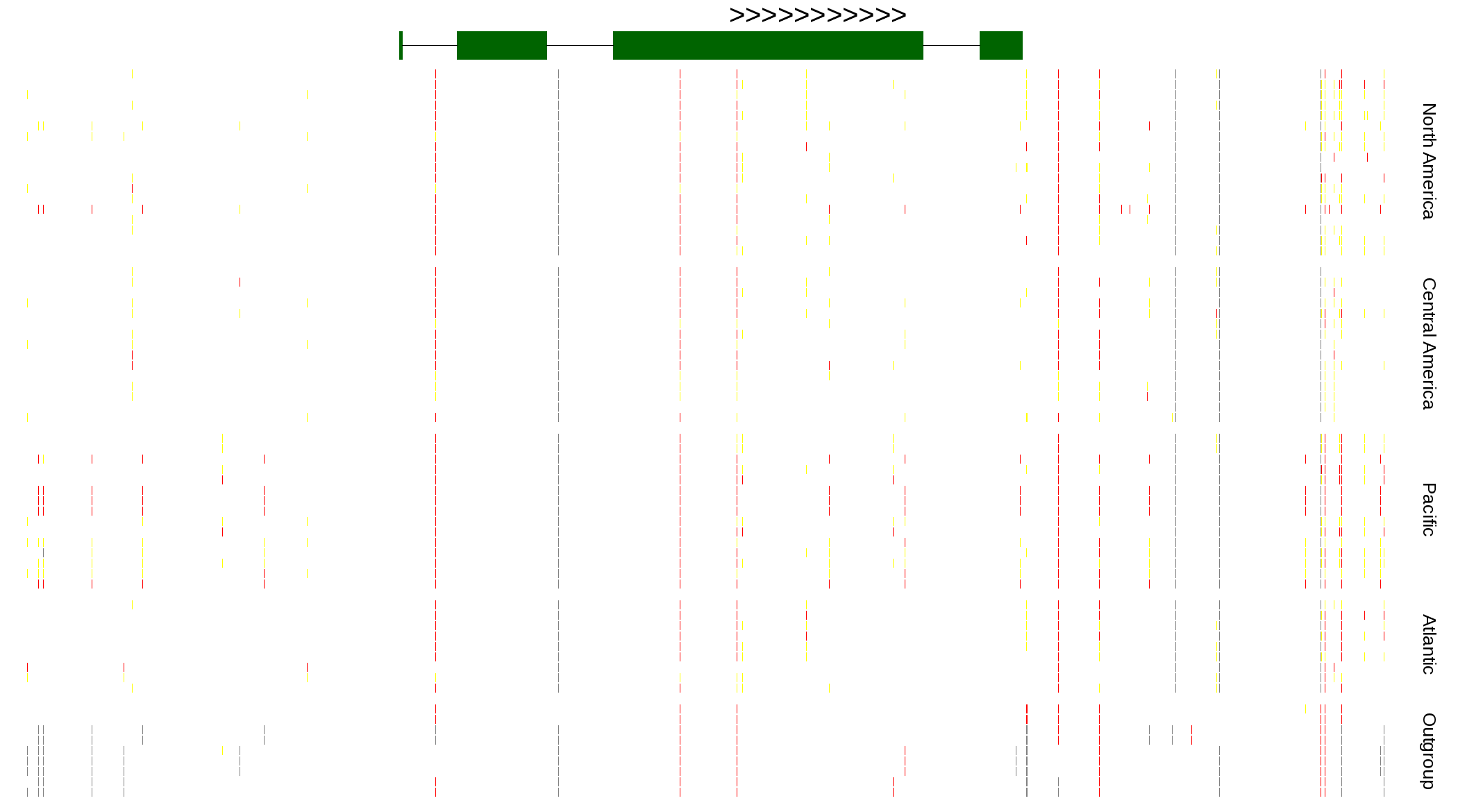
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS208935-TA
ATGATCTCGAAACCAGCCCCGGAGTGGGAGGCTACAGCTGTCGTTAATGGCGAGATAACGCAACTTTCACTATCAAGTTTTAAAGGAAAATATTTAGTCTTCTTCTTTTATCCCTTAGACTTCACCTTTGTTTGTCCTACTGAAATTTTGGCCTTCTCTGAAAGAATTGAAGAGTTCAGAAAACTCAACACTGAAGTAGTAGCATGTTCCGTAGACTCACACTTTACTCATCTAGCCTGGATTAACACTCCTCGGAAAGAGGGAGGTCTAGGAAAAATTAAAATCCCATTATTAAGTGATCTCACACATTCCATTGCCAAAGACTATGGGGTATATTTAGAAGATTTGGGTCACACTTTGAGAGGTTTATTCATAATTGATGATAAAGGAATTCTTAGACAAATTACAATGAATGATCTGCCAGTTGGCAGATCTGTGGATGAAACGCTAAGGCTGGTTCAAGCATTCCAGTACACTGACAAACATGGCGAGGTTTGTCCGGCGGGATGGAAACCAGGTCAAGATACAATTATTCCCAATCCTGATGAAAAGAAAAAATATTTTGAAAAGTTGTCAGATCTATAG
>DPOGS208935-PA
MISKPAPEWEATAVVNGEITQLSLSSFKGKYLVFFFYPLDFTFVCPTEILAFSERIEEFRKLNTEVVACSVDSHFTHLAWINTPRKEGGLGKIKIPLLSDLTHSIAKDYGVYLEDLGHTLRGLFIIDDKGILRQITMNDLPVGRSVDETLRLVQAFQYTDKHGEVCPAGWKPGQDTIIPNPDEKKKYFEKLSDL-