| DPOGS210365 | ||
|---|---|---|
| Transcript | DPOGS210365-TA | 1131 bp |
| Protein | DPOGS210365-PA | 376 aa |
| Genomic position | DPSCF300025 + 471670-472800 | |
| RNAseq coverage | 2311x (Rank: top 5%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL013834 | 0.0 | 92.55% | |
| Bombyx | BGIBMGA011922-TA | 0.0 | 91.03% | |
| Drosophila | Fdh-PA | 8e-168 | 72.94% | |
| EBI UniRef50 | UniRef50_P80467 | 2e-171 | 75.34% | Alcohol dehydrogenase class-3 n=114 Tax=cellular organisms RepID=ADHX_UROHA |
| NCBI RefSeq | NP_001040507.1 | 0.0 | 92.02% | alcohol dehydrogenase [Bombyx mori] |
| NCBI nr blastp | gi|114052488 | 0.0 | 92.02% | alcohol dehydrogenase [Bombyx mori] |
| NCBI nr blastx | gi|114052488 | 0.0 | 92.27% | alcohol dehydrogenase [Bombyx mori] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008270 | 7.9e-215 | zinc ion binding | |
| GO:0055114 | 7.9e-215 | oxidation-reduction process | ||
| GO:0016491 | 7.9e-215 | oxidoreductase activity | ||
| GO:0006069 | 5.9e-195 | ethanol oxidation | ||
| GO:0051903 | 5.9e-195 | S-(hydroxymethyl)glutathione dehydrogenase activity | ||
| GO:0005488 | 3.3e-36 | binding | ||
| KEGG pathway | dre:116517 | 0.0 | ||
| K00121 (frmA, ADH5, adhC) | maps-> | Drug metabolism - cytochrome P450 | ||
| Glycolysis / Gluconeogenesis | ||||
| Fatty acid metabolism | ||||
| 3-Chloroacrylic acid degradation | ||||
| Tyrosine metabolism | ||||
| Metabolism of xenobiotics by cytochrome P450 | ||||
| Methane metabolism | ||||
| 1- and 2-Methylnaphthalene degradation | ||||
| Retinol metabolism | ||||
| InterPro domain | [1-377] IPR002085 | 7.9e-215 | Alcohol dehydrogenase superfamily, zinc-type | |
| [9-376] IPR014183 | 5.9e-195 | Alcohol dehydrogenase class III/S-(hydroxymethyl)glutathione dehydrogenase | ||
| [2-200] IPR011032 | 3.4e-84 | GroES-like | ||
| [197-316] IPR016040 | 3.3e-36 | NAD(P)-binding domain | ||
| [35-160] IPR013154 | 4.5e-25 | Alcohol dehydrogenase GroES-like | ||
| [204-326] IPR013149 | 4.2e-21 | Alcohol dehydrogenase, C-terminal | ||
| Orthology group | MCL11155 | Single-copy universal gene | ||
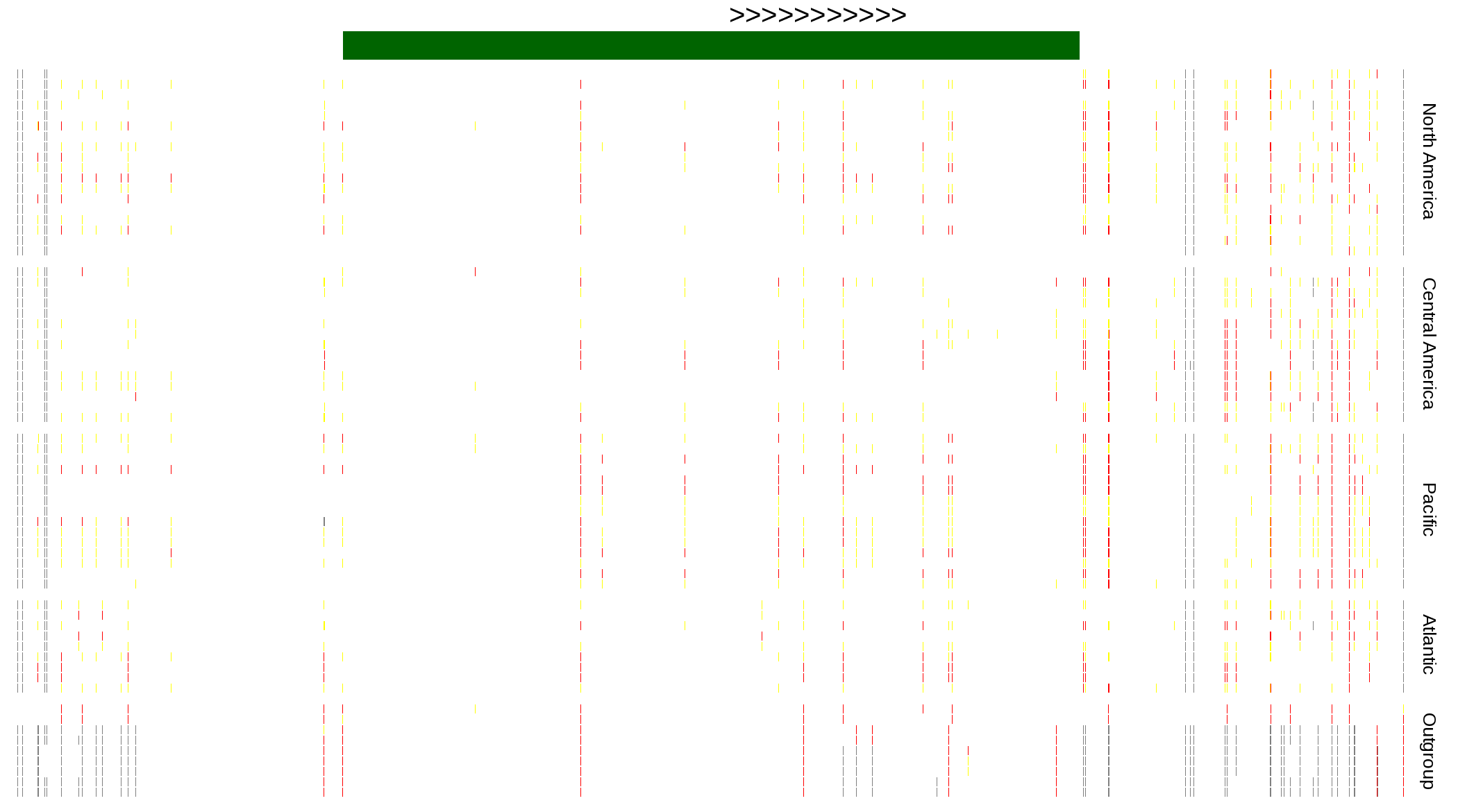
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS210365-TA
ATGTCTACTGCTGGAAAAGTTATTAAATGTAAAGCAGCTGTAGCTTGGGAAGCTGGAAAGCCATTATCTATTGAAGAAATCGAAGTTGATCCACCAAAAGCTGGAGAAGTACGGGTAAAAATTCTTGCTACCGGTGTCTGTCACACAGATGCATATACTTTGTCCGGAAAAGATCCAGAAGGAGTCTTTCCAGTCATTTTAGGGCATGAAGGCGGTGGTGTAGTCGAAAGTGTTGGCGAGGGAGTGACGTCCGTGAAGCCCGGCGATCATGTGTTACCTCTGTACGTCCCGCAGTGCAAAACATGCAAATTCTGTCAAAACCCTAAAACCAACCTTTGTCAGAAAGTGAGAATCACGCAGGGACAGGGTGTCATGCCTGATGGCACGAAGAGATTCCGATGCAAAGGTCAAGAGCTGTTCCACTTCATGGGGTGCTCAACGTTCAGTGAATATACTGTAGTTCTAGAAATTTCGTTGTGCAAGATAGCTGACGCCGCGCCTCTTGATAAGGTATGTCTATTGGGATGTGGTGTGACAACCGGCTATGGAGCCGCTCTCAATACAGCAAAGGTGGAACCGGGTTCGAACTGCGCTATATTCGGTCTCGGAGCTGTCGGCCTGGCCGTAGCTCTCGGTTGTAAAGCAGCGGGGGCAAATCGCATCATAGGAGTCGACATTAACCCTGACAAATATGAGATTGCCAAAAAATTTGGCGTCAATGAATTCGTCAATCCCAAGGACTACGACAAACCTATCCAGCAGGTACTTGTTGACTTGACAGATGGAGGATTGGAGTATACATTTGAGTGCATAGGTAATGTTAATACTATGCGTGCTGCCCTGGAGGCATGCCATAAGGGTTGGGGTGAGTCTGTTATCATTGGTGTGGCCGCCGCCGGGGAAGAGATCAGCACCCGTCCATTCCAACTCGTGACCGGCCGAGTATGGAGAGGAACAGCCTTCGGAGGCTACAAGAGCCGTGACAGTGTTCCTAAGTTGGTTGATGACTACCTCAATAAGAAACTTCCTTTGGATGACTTCGTTACTCATAAAGTACCTCTCAGTGAGATTAACGAGGCCTTCCATTTAATGCACGTGGGGAAATCCATCCGTGCCGTTGTTGAGTTTTAA
>DPOGS210365-PA
MSTAGKVIKCKAAVAWEAGKPLSIEEIEVDPPKAGEVRVKILATGVCHTDAYTLSGKDPEGVFPVILGHEGGGVVESVGEGVTSVKPGDHVLPLYVPQCKTCKFCQNPKTNLCQKVRITQGQGVMPDGTKRFRCKGQELFHFMGCSTFSEYTVVLEISLCKIADAAPLDKVCLLGCGVTTGYGAALNTAKVEPGSNCAIFGLGAVGLAVALGCKAAGANRIIGVDINPDKYEIAKKFGVNEFVNPKDYDKPIQQVLVDLTDGGLEYTFECIGNVNTMRAALEACHKGWGESVIIGVAAAGEEISTRPFQLVTGRVWRGTAFGGYKSRDSVPKLVDDYLNKKLPLDDFVTHKVPLSEINEAFHLMHVGKSIRAVVEF-