| DPOGS210783 | ||
|---|---|---|
| Transcript | DPOGS210783-TA | 198 bp |
| Protein | DPOGS210783-PA | 65 aa |
| Genomic position | DPSCF301335 + 3065-3486 | |
| RNAseq coverage | 1524x (Rank: top 8%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL014821 | 2e-15 | 56.72% | |
| Bombyx | BGIBMGA014453-TA | 5e-19 | 73.58% | |
| Drosophila | CG6084-PB | 2e-15 | 61.19% | |
| EBI UniRef50 | UniRef50_G3MKM4 | 5e-12 | 51.52% | Putative uncharacterized protein n=2 Tax=Amblyomma maculatum RepID=G3MKM4_9ACAR |
| NCBI RefSeq | XP_001944024.1 | 6e-18 | 63.64% | PREDICTED: similar to AGAP011050-PA [Acyrthosiphon pisum] |
| NCBI nr blastp | gi|193601268 | 1e-16 | 63.64% | PREDICTED: aldo-keto reductase family 1 member B10-like [Acyrthosiphon pisum] |
| NCBI nr blastx | gi|193601268 | 5e-16 | 63.64% | PREDICTED: aldo-keto reductase family 1 member B10-like [Acyrthosiphon pisum] |
| Group | ||||
|---|---|---|---|---|
| KEGG pathway | dwi:Dwil_GK17015 | 4e-17 | ||
| K00011 (E1.1.1.21, AKR1) | maps-> | Galactose metabolism | ||
| Glycerolipid metabolism | ||||
| Pentose and glucuronate interconversions | ||||
| Fructose and mannose metabolism | ||||
| Pyruvate metabolism | ||||
| InterPro domain | [1-65] IPR001395 | 3.7e-22 | Aldo/keto reductase | |
| [1-64] IPR023210 | 4.2e-14 | NADP-dependent oxidoreductase domain | ||
| Orthology group | ||||
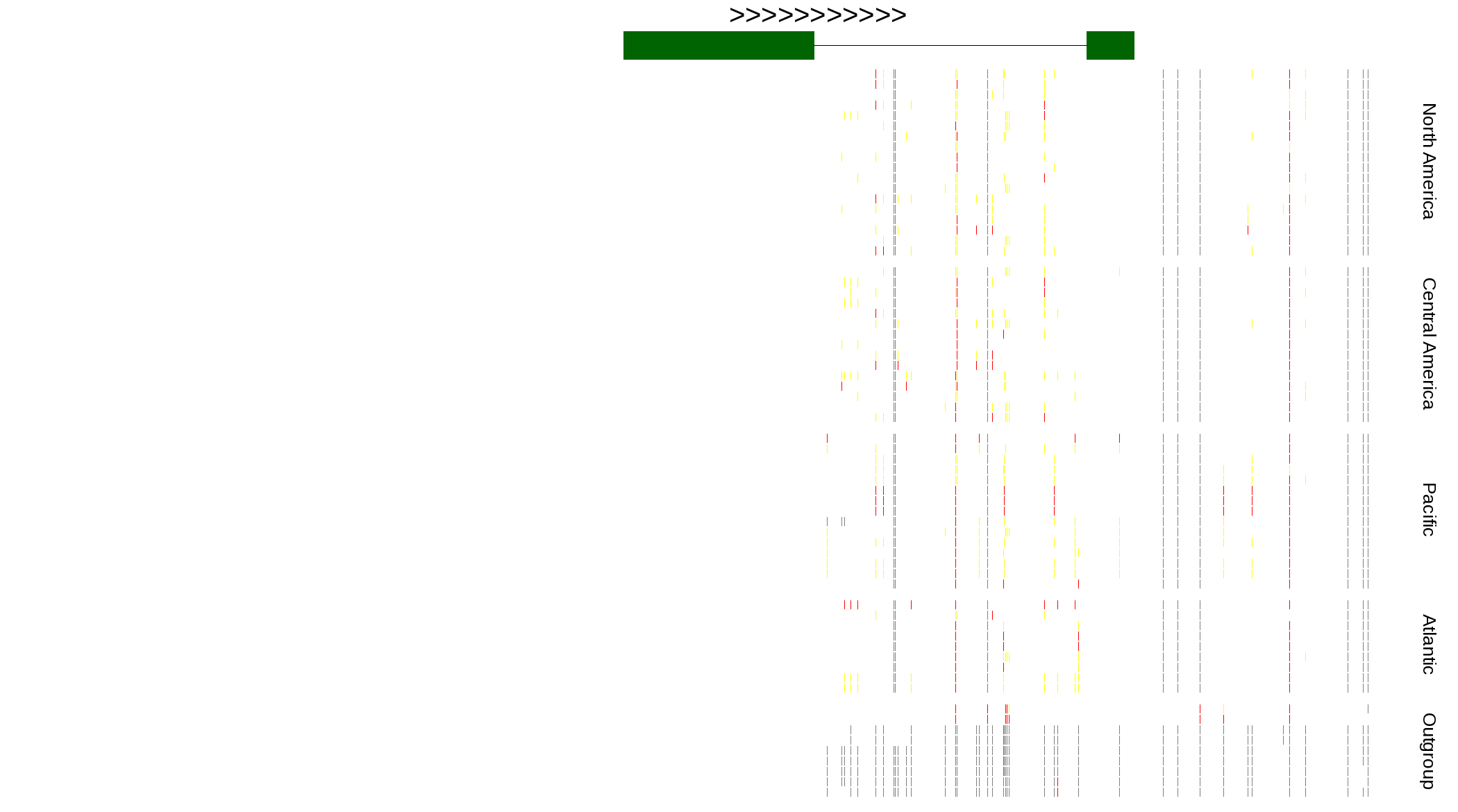
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS210783-TA
AGGTACGCGATCGACCGAGGCCTAATAGTGATCCCCAAATCGGTGACGAAGAGTCGCATCGAACAGAACTTCCAGGTGCTGGATTTCCAGCTTTCACCTGAAGACATTGACCTCATCGCTACATTTGAATGTAACGGACGACTCTGTACTATGGGAGATGTCCACCATCCGGACTATCCATTCCACGATGAATATTAA
>DPOGS210783-PA
RYAIDRGLIVIPKSVTKSRIEQNFQVLDFQLSPEDIDLIATFECNGRLCTMGDVHHPDYPFHDEY-