| DPOGS211805 | ||
|---|---|---|
| Transcript | DPOGS211805-TA | 693 bp |
| Protein | DPOGS211805-PA | 230 aa |
| Genomic position | DPSCF300031 - 698275-699790 | |
| RNAseq coverage | 137x (Rank: top 55%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL022141 | 2e-30 | 35.75% | |
| Bombyx | BGIBMGA005131-TA | 5e-34 | 38.99% | |
| Drosophila | Cp1-PC | 2e-34 | 37.69% | |
| EBI UniRef50 | UniRef50_D8R100 | 7e-40 | 43.64% | Putative uncharacterized protein n=4 Tax=Selaginella moellendorffii RepID=D8R100_SELML |
| NCBI RefSeq | XP_002161512.1 | 8e-38 | 39.17% | PREDICTED: similar to Cathepsin L [Hydra magnipapillata] |
| NCBI nr blastp | gi|302790570 | 9e-40 | 43.64% | hypothetical protein SELMODRAFT_268054 [Selaginella moellendorffii] |
| NCBI nr blastx | gi|302790570 | 3e-40 | 43.64% | hypothetical protein SELMODRAFT_268054 [Selaginella moellendorffii] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008234 | 3.9e-72 | cysteine-type peptidase activity | |
| GO:0006508 | 1.2e-70 | proteolysis | ||
| KEGG pathway | spu:575142 | 2e-37 | ||
| K01365 (CTSL) | maps-> | Lysosome | ||
| Phagosome | ||||
| Antigen processing and presentation | ||||
| InterPro domain | [20-220] IPR013128 | 3.9e-72 | Peptidase C1A, papain | |
| [19-228] IPR000668 | 1.2e-70 | Peptidase C1A, papain C-terminal | ||
| Orthology group | MCL27835 | Specific divergent | ||
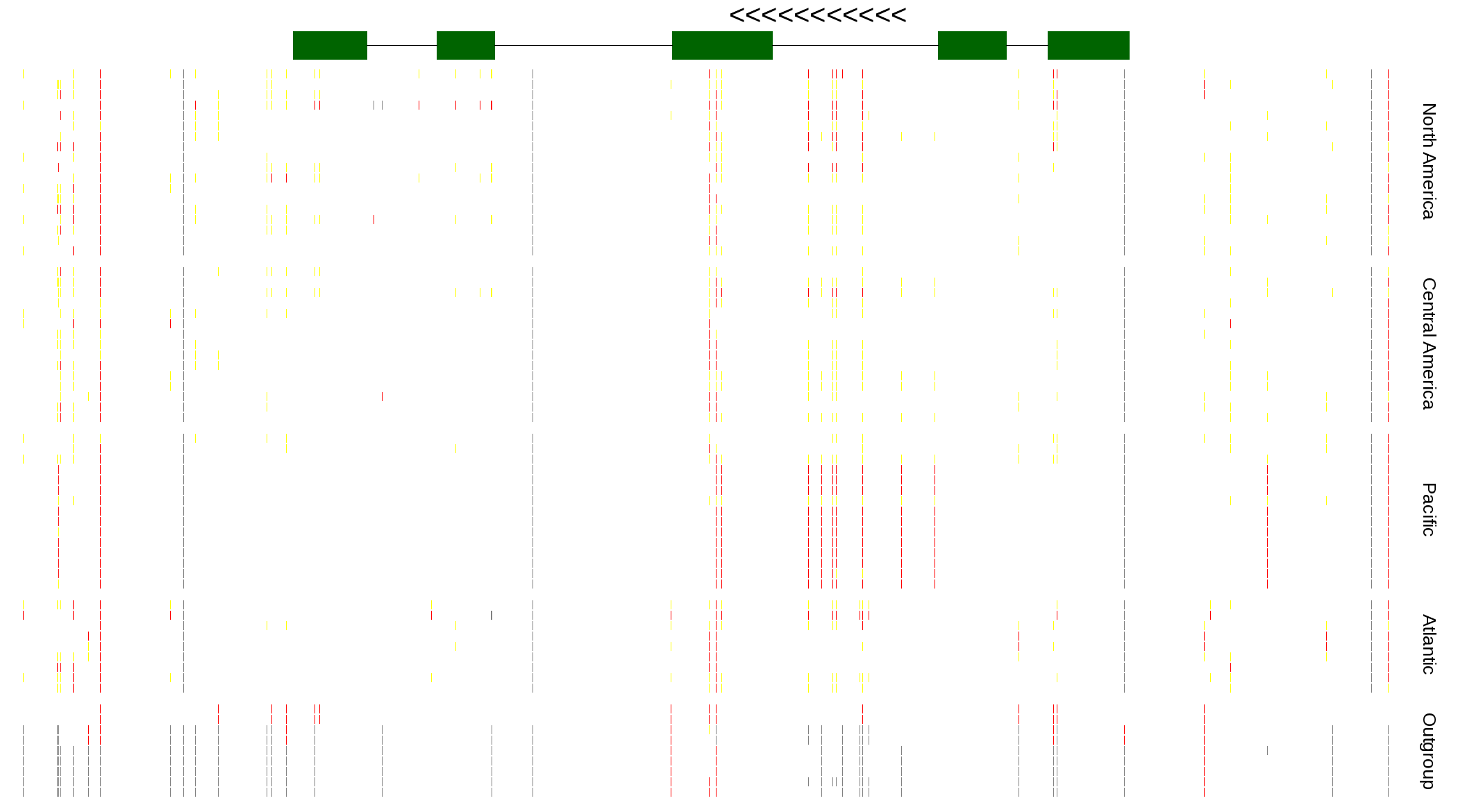
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS211805-TA
ATGGGTTCACATATTGATGTCACAAGTTTGAAGAAATACGAGCCAAAAGGATTTGCACCAGCGAGCTTGGACTATCGTAGCTATGGTTGGGTGACTGAAATAAAAGATCAAAGACACTGTAATACATGTTATATTTTCAGCGCGATTGGCGCTATCGAAGGACAGCACGCAAAACTTCATGGAAACCTAGTTTCTTTATCAGAACAGCAAGAATTGGATTGTGAGAGCGGGGATGGTTGCAAGACGGGAGGATTTCCTCATGAAGCTATGAATGTTCTCGCTCAGCAAGGAGGCTCTATGTCTGAAGAAAAGTATCCGTATGAGGAAGGAAAGGGACAGTGCCGTACTGATAAATCCAAGATTGTTGTTAAAGTAACCGGAGGATCACAGTTGACAGTTTCCAGCGAAGATGACTTAAAAGACGCTTTGGCAAACAACGGCCCACTGTCCATCGCTTTATTTATTTGTCGGGAATTCCAACATTATACTGGAGGCATATTTGTTCACAACTGCCAGGGAGATGACGGGCATGCTGTGGTACTTGTAGGCTACGATTCAGCTGATGGCCAGGAATACTGGATCATCAAGAACTCGTGGGCCACGGTCTGGGGAGAGCAAGGATACATGCGAATGAAGTTAGGATCATCCCTTTGCCAAATAGGATATTATATCGCTCTGGCTTCAGTAGCATAG
>DPOGS211805-PA
MGSHIDVTSLKKYEPKGFAPASLDYRSYGWVTEIKDQRHCNTCYIFSAIGAIEGQHAKLHGNLVSLSEQQELDCESGDGCKTGGFPHEAMNVLAQQGGSMSEEKYPYEEGKGQCRTDKSKIVVKVTGGSQLTVSSEDDLKDALANNGPLSIALFICREFQHYTGGIFVHNCQGDDGHAVVLVGYDSADGQEYWIIKNSWATVWGEQGYMRMKLGSSLCQIGYYIALASVA-