| DPOGS212790 | ||
|---|---|---|
| Transcript | DPOGS212790-TA | 651 bp |
| Protein | DPOGS212790-PA | 216 aa |
| Genomic position | DPSCF300489 - 56351-58939 | |
| RNAseq coverage | 0x (Rank: top 99%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL011174 | 6e-14 | 66.04% | |
| Bombyx | BGIBMGA009631-TA | 9e-11 | 54.39% | |
| Drosophila | % | |||
| EBI UniRef50 | UniRef50_P43513 | 3e-11 | 43.70% | Chorion class A protein Ld3/Ld29 n=16 Tax=Obtectomera RepID=CHA3_LYMDI |
| NCBI RefSeq | NP_001112376.1 | 5e-06 | 40.35% | chorion class CA protein ERA.1 precursor [Bombyx mori] |
| NCBI nr blastp | gi|1168930 | 1e-14 | 46.15% | chorion protein [Lymantria dispar] |
| NCBI nr blastx | gi|1168930 | 3e-17 | 46.15% | chorion protein [Lymantria dispar] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0007304 | 1.4e-10 | chorion-containing eggshell formation | |
| GO:0005213 | 1.4e-10 | structural constituent of chorion | ||
| GO:0007275 | 1.4e-10 | multicellular organismal development | ||
| GO:0042600 | 1.4e-10 | chorion | ||
| KEGG pathway | ||||
| InterPro domain | [5-140] IPR002635 | 1.4e-10 | Chorion protein | |
| Orthology group | MCL10869 | Lepidoptera specific | ||
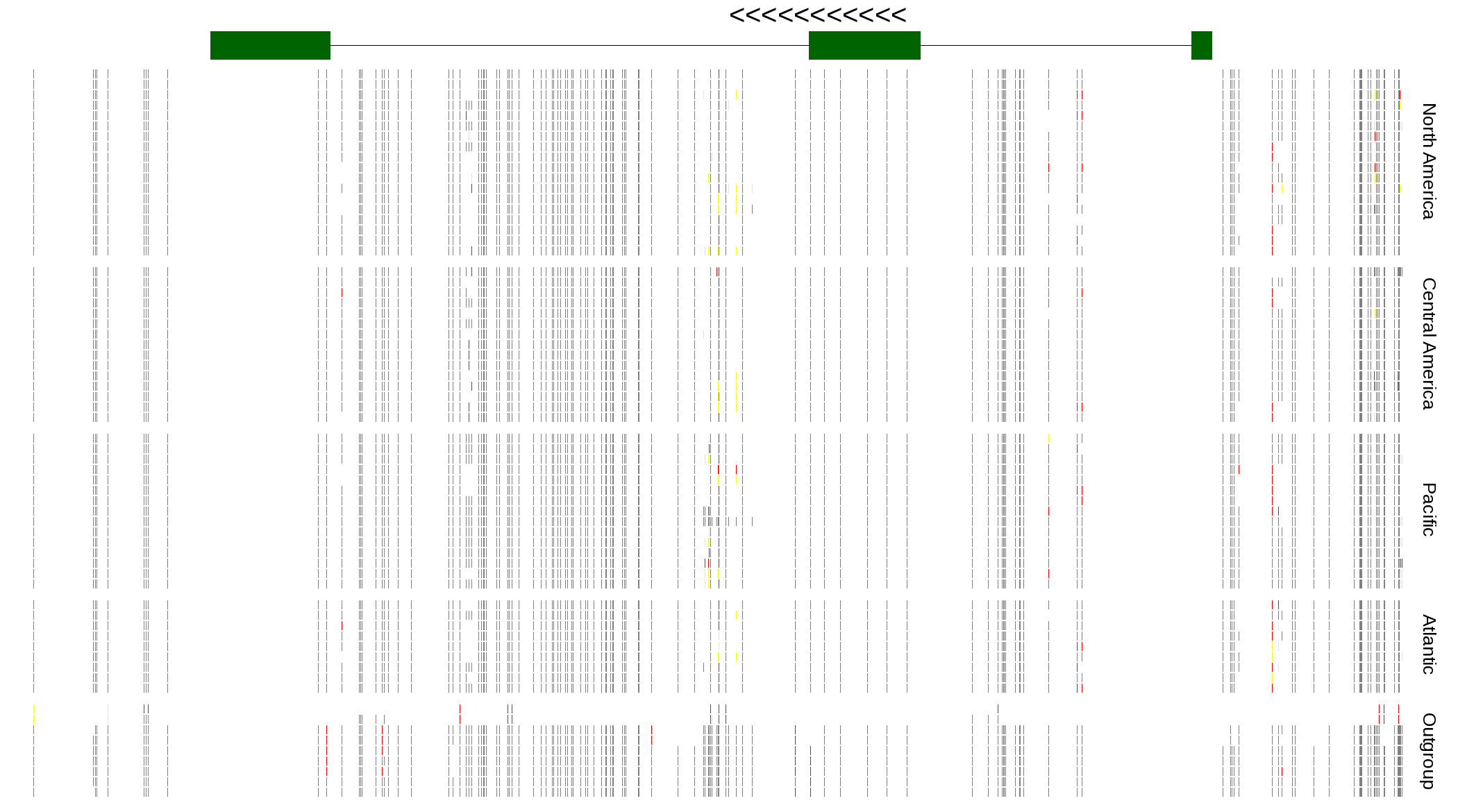
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS212790-TA
ATGTTTGTTCCATTTGCATTCCTTTTTGTCTGCATCCAAGCTTGTTTGGTTCAGGATGTATACAGTCAATGTCTTAACAGGGCTGCGCTCGGAACTCCACTTGGACAAGGTTTTGTATCAGGAGTAGGACTTGGCTCGTGTGCTTCTGAAACTTATGGGTCACTCGGACTGAACTCTCAAGTGGACTTGGTCGAGGAACAAGGAGCGAGTTACGGTGGAACCGGGGTCGGAGAGCTTTATGTGAACGGATTGTTACCGGTGGAAGGATATTCATCCATCAGTGGACAGGTCCCCGTCCTTGGCGTAGTGAGATTTGACGGACCAGTAGCGGCCAGGGGGAGTAACGCTTACAGCACGTGTATCGGTGCTGGTTTGTCTGGTTTTGCTCCTCGTTATGGTCCACTCGGCTTCACGCCAGACATGTCGCCTGTTGTCGATCCAACGTCATATTCTCAAGCATACGGTGGAGAAGGTCTCGGTGACATCAGTATTGTTGGAAAGATGCCGGTCTCTGGAATCACAGCGGTGAAGGGCCAGGTCCCAATTCTTGGTACAGTAAACTTTGGGGGTGATGTAAACGCCGCTGGCACGGTTACCATCACTGGTAGCAGTGGATACAGAGCTTACAGAGGTTACAGAGATTACTGCTAA
>DPOGS212790-PA
MFVPFAFLFVCIQACLVQDVYSQCLNRAALGTPLGQGFVSGVGLGSCASETYGSLGLNSQVDLVEEQGASYGGTGVGELYVNGLLPVEGYSSISGQVPVLGVVRFDGPVAARGSNAYSTCIGAGLSGFAPRYGPLGFTPDMSPVVDPTSYSQAYGGEGLGDISIVGKMPVSGITAVKGQVPILGTVNFGGDVNAAGTVTITGSSGYRAYRGYRDYC-