| DPOGS213171 | ||
|---|---|---|
| Transcript | DPOGS213171-TA | 918 bp |
| Protein | DPOGS213171-PA | 305 aa |
| Genomic position | DPSCF300114 - 331554-334239 | |
| RNAseq coverage | 649x (Rank: top 20%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL002989 | 1e-121 | 67.96% | |
| Bombyx | BGIBMGA007360-TA | 5e-144 | 80.07% | |
| Drosophila | CG4389-PA | 9e-35 | 31.77% | |
| EBI UniRef50 | UniRef50_E0VKS4 | 3e-100 | 59.12% | Short chain 3-hydroxyacyl-CoA dehydrogenase, putative n=10 Tax=Opisthokonta RepID=E0VKS4_PEDHC |
| NCBI RefSeq | NP_001040414.1 | 1e-135 | 77.12% | 3-hydroxyacyl-CoA dehydrogenase [Bombyx mori] |
| NCBI nr blastp | gi|114050917 | 3e-134 | 77.12% | 3-hydroxyacyl-CoA dehydrogenase [Bombyx mori] |
| NCBI nr blastx | gi|114050917 | 6e-129 | 77.12% | 3-hydroxyacyl-CoA dehydrogenase [Bombyx mori] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0006631 | 7.3e-57 | fatty acid metabolic process | |
| GO:0003857 | 7.3e-57 | 3-hydroxyacyl-CoA dehydrogenase activity | ||
| GO:0055114 | 7.3e-57 | oxidation-reduction process | ||
| GO:0016491 | 7.3e-57 | oxidoreductase activity | ||
| GO:0005488 | 1.9e-54 | binding | ||
| GO:0016616 | 2.3e-33 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | ||
| GO:0050662 | 2.3e-33 | coenzyme binding | ||
| KEGG pathway | tca:661810 | 5e-105 | ||
| K00022 (HADH) | maps-> | Fatty acid elongation in mitochondria | ||
| Tryptophan metabolism | ||||
| Lysine degradation | ||||
| Valine, leucine and isoleucine degradation | ||||
| Geraniol degradation | ||||
| Fatty acid metabolism | ||||
| Caprolactam degradation | ||||
| Butanoate metabolism | ||||
| InterPro domain | [22-205] IPR006176 | 7.3e-57 | 3-hydroxyacyl-CoA dehydrogenase, NAD binding | |
| [19-208] IPR016040 | 1.9e-54 | NAD(P)-binding domain | ||
| [207-303] IPR006108 | 1.4e-34 | 3-hydroxyacyl-CoA dehydrogenase, C-terminal | ||
| [209-304] IPR013328 | 2.3e-33 | Dehydrogenase, multihelical | ||
| [207-304] IPR008927 | 7.2e-32 | 6-phosphogluconate dehydrogenase, C-terminal-like | ||
| Orthology group | MCL17816 | Patchy | ||
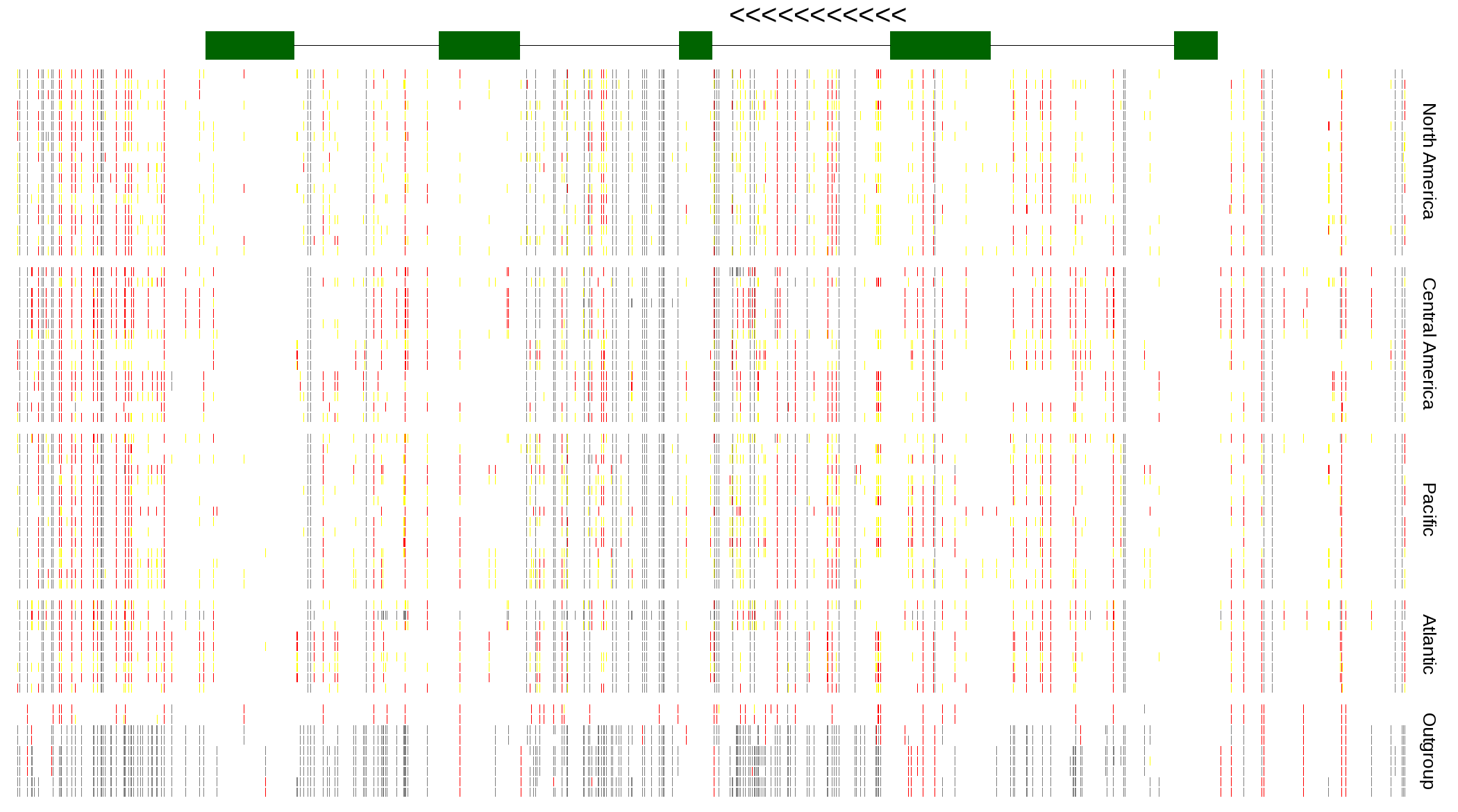
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS213171-TA
ATGAATAATTTGAACGTTATATGTAGAAAGTTTTCCCATTCGGCATCTTTAAATGCTATCAAAACGGTAACAGTTGTCGGCGGCGGGCTTATGGGTTCTGGAATCGCACAGGCAACTCAAGCCGGACAAAATGTAACAATAGTCGATTTGAACTCGGAAATACTTGAAAAAGCTCAGAAATCTATACAAAACAATCTAGGCAGAGTGGCGAGAAAGCTGTATAAAGATGATCCTTTGAAAATGGAGGAATTTGTTAAAGAAGCCAATAGTAGAATTAAGGTTTCAACCAAGATTGAAGATGGTGTGGACGCTGATCTGATCGTAGAGGCCATCGTCGAGTTACTTGAGCCCAAACAGAAGCTGTTCAACAGATTGGATGAGCTGGCTCCAGAACATACAATTTTAGCGAGCAACACATCATCTATATCAATCAATGAGATAGGCAGCGGTATTAAGAGAAAAGATAGGTTTGGTGGGCTGCACTTCTTCAATCCTGTGCCTGTGATGCGTCTCCTGGAGGTTATCAAGAGCGACCGCATGTCCCAAGAGACTTATAACGCCATGATGGAGTGGGGCAAGTCCGTGGGCAAGACCTGCATCACCTGCAAAGATACTCCTGGATTTGTCGTCAACAGGCTACTAGGGCCTTACAGTGCTGAAGCTTTCAGAATGTTTGAGCGAGGTGATGCCAGTAAAGAAGACATCGATATTGCTATGAAGTTGGGTGCGGGATACCCCATGGGTCCGCTAGAGCTGGCCGACTACACCGGACTCGATACTAACAAGTTCGTCCTCGAGGTGTTGTACCAGAAGACAAAGAACCAAGTGTTTAAGCCGATACCGTTACTGAATAAAATGGTGGAAGAAGGCAAACTGGGAATCAAGACCGGGGAGGGGATCTATAAATACAAGAAGTGA
>DPOGS213171-PA
MNNLNVICRKFSHSASLNAIKTVTVVGGGLMGSGIAQATQAGQNVTIVDLNSEILEKAQKSIQNNLGRVARKLYKDDPLKMEEFVKEANSRIKVSTKIEDGVDADLIVEAIVELLEPKQKLFNRLDELAPEHTILASNTSSISINEIGSGIKRKDRFGGLHFFNPVPVMRLLEVIKSDRMSQETYNAMMEWGKSVGKTCITCKDTPGFVVNRLLGPYSAEAFRMFERGDASKEDIDIAMKLGAGYPMGPLELADYTGLDTNKFVLEVLYQKTKNQVFKPIPLLNKMVEEGKLGIKTGEGIYKYKK-